Regulation of co-translational mRNA decay by PAP and DXO1 in Arabidopsis
- PMID: 39966730
- PMCID: PMC11834196
- DOI: 10.1186/s12870-025-06195-5
Regulation of co-translational mRNA decay by PAP and DXO1 in Arabidopsis
Abstract
Background: mRNA decay is central in the regulation of mRNA homeostasis in the cell. The recent discovery of a co-translational mRNA decay pathway (also called CTRD) has changed our understanding of the mRNA decay process. This pathway has emerged as an evolutionarily conversed mechanism essential for specific physiological processes in eukaryotes, especially in plants. In Arabidopsis, this pathway is targeted mainly by the exoribonuclease XRN4. However, the details of the molecular regulation of this pathway are still unclear.
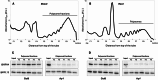
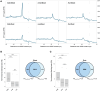
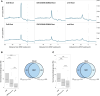
Results: In this study, we first tested the role of the 3'-phosphoadenosine 5'-phosphate (PAP), an inhibitor of exoribonucleases in the regulation of CTRD. Using 5'Pseq approach, we discovered that FRY1 inactivation impaired XRN4-CTRD activity. Based on this finding, we demonstrated that exogenous PAP treatment stabilizes CTRD mRNA targets. Furthermore, we also tested the implication of the exoribonuclease DXO1 in CTRD regulation. We found that DXO1, another exoribonuclease sensitive to PAP, is also involved in the CTRD pathway, probably by targeting NAD+-capped mRNAs. DXO1 specifically targets mRNAs linked to stress response.
Conclusions: Our study provides further insights into the regulation of CTRD in Arabidopsis and demonstrates that other exoribonucleases can be implicated in this pathway.
Keywords: 3ʹ-phosphoadenosine 5ʹ-phosphate; Arabidopsis; Co-translational mRNA decay; DXO1; FRY1; XRN4.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not Applicable. Consent for publication: Not Applicable. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Genome-wide analysis of mRNA decay in Arabidopsis shoot and root reveals the importance of co-translational mRNA decay in the general mRNA turnover.Nucleic Acids Res. 2024 Jul 22;52(13):7910-7924. doi: 10.1093/nar/gkae363. Nucleic Acids Res. 2024. PMID: 38721772 Free PMC article.
-
Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories.RNA. 2011 Mar;17(3):501-11. doi: 10.1261/rna.2467911. Epub 2011 Jan 11. RNA. 2011. PMID: 21224377 Free PMC article.
-
Mechanistic insights into DXO1 and XRN3: regulatory roles of RNA stability, transcription, and liquid-liquid phase separation in Arabidopsis thaliana (L.) Heynh.Plant Sci. 2025 Apr;353:112413. doi: 10.1016/j.plantsci.2025.112413. Epub 2025 Feb 3. Plant Sci. 2025. PMID: 39909287 Review.
-
Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors.Plant Cell. 2007 Nov;19(11):3451-61. doi: 10.1105/tpc.107.055319. Epub 2007 Nov 9. Plant Cell. 2007. PMID: 17993620 Free PMC article.
-
Activity and roles of Arabidopsis thaliana XRN family exoribonucleases in noncoding RNA pathways.J Plant Res. 2017 Jan;130(1):25-31. doi: 10.1007/s10265-016-0887-z. Epub 2016 Dec 17. J Plant Res. 2017. PMID: 27988817 Review.
Cited by
-
RNA interference and turnover in plants -a complex partnership.Front Plant Sci. 2025 Jul 1;16:1608888. doi: 10.3389/fpls.2025.1608888. eCollection 2025. Front Plant Sci. 2025. PMID: 40666303 Free PMC article. Review.
References
-
- Belostotsky DA, Sieburth LE. Kill the messenger: mRNA decay and plant development. Curr Opin Plant Biol [Internet]. 2009;12(1):96–102. Available from: https://www.sciencedirect.com/science/article/pii/S1369526608001581 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

