Immunomodulation by allograft endothelial cells
- PMID: 39967861
- PMCID: PMC11832486
- DOI: 10.3389/frtra.2025.1518772
Immunomodulation by allograft endothelial cells
Abstract
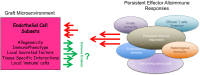
It is increasingly appreciated that the expression of immunoregulatory molecules within tumors have potential to shape a microenvironment that promotes local immunoevasion and immunoregulation. However, little is known about tissue-intrinsic immunomodulatory mechanisms following transplantation. We propose that differences in the phenotype of microvascular endothelial cells impact the alloantigenicity of the graft and its potential to promote immunoregulation following transplantation. We focus this review on the concept that graft-dependent immunoregulation may evolve post-transplantation, and that it is dependent on the phenotype of select subsets of intragraft endothelial cells. We also discuss evidence that long-term graft survival is critically dependent on adaptive interactions among immune cells and endothelial cells within the transplanted tissue microenvironment.
Keywords: allograft (ALLO); endothelial cell; graft survival; immunoregulation; transplantation.
© 2025 Bose, Do, Testini, Jadhav, Sailliet, Kho, Komatsu, Boneschansker, Kong, Wedel and Briscoe.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Ingulli E, Alexander SI, Briscoe DM. Immunology of pediatric renal transplantation. In: Avner ED, Harmon WE, Niaudet P, Yoshikawa N, Emma F, Goldstein SL, editors. Pediatric Nephrology. New York, NY: Springer; (2016). p. 2457–500.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

