Early-life antibiotic exposure aggravates hepatic steatosis through enhanced endotoxemia and lipotoxic effects driven by gut Parabacteroides
- PMID: 39968496
- PMCID: PMC11832435
- DOI: 10.1002/mco2.70104
Early-life antibiotic exposure aggravates hepatic steatosis through enhanced endotoxemia and lipotoxic effects driven by gut Parabacteroides
Abstract
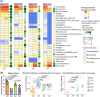
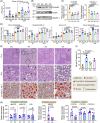
Compelling evidence supports a link between early-life gut microbiota and the metabolic outcomes in later life. Using an early-life antibiotic exposure model in BALB/c mice, we investigated the life-course impact of prenatal and/or postnatal antibiotic exposures on the gut microbiome of offspring and the development of metabolic dysfunction-associated steatotic liver disease (MASLD). Compared to prenatal antibiotic exposure alone, postnatal antibiotic exposure more profoundly affected gut microbiota development and succession, which led to aggravated endotoxemia and metabolic dysfunctions. This was primarily resulted from the overblooming of gut Parabacteroides and hepatic accumulation of cytotoxic lysophosphatidyl cholines (LPCs), which acted in conjunction with LPS derived from Parabacteroides distasonis (LPS_PA) to induce cholesterol metabolic dysregulations, endoplasmic reticulum (ER) stress and apoptosis. Integrated serum metabolomics, hepatic lipidomics and transcriptomics revealed enhanced glycerophospholipid hydrolysis and LPC production in association with the upregulation of PLA2G10, the gene controlling the expression of the group X secretory Phospholipase A2s (sPLA2-X). Taken together, our results show microbial modulations on the systemic MASLD pathogenesis and hepatocellular lipotoxicity pathways following early-life antibiotic exposure, hence help inform refined clinical practices to avoid any prolonged maternal antibiotic administration in early life and potential gut microbiota-targeted intervention strategies.
Keywords: antibiotics; early life; gut microbiota; lipotoxicity; metabolic dysfunction‐associated steatotic liver disease.
© 2025 The Author(s). MedComm published by Sichuan International Medical Exchange & Promotion Association (SCIMEA) and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
F.K.L.C. is Board Member of CUHK Medical Centre. He is a cofounder, nonexecutive Board Chairman and shareholder of GenieBiome Ltd. He receives patent royalties through his affiliated institutions. He has received fees as an advisor and honoraria as a speaker for Eisai Co. Ltd., AstraZeneca, Pfizer Inc., Takeda Pharmaceutical Co., and Takeda (China) Holdings Co. Ltd. S.C.N. has served as an advisory board member for Pfizer, Ferring, Janssen, and Abbvie and received honoraria as a speaker for Ferring, Tillotts, Menarini, Janssen, Abbvie, and Takeda. S.C.N. has received research grants through her affiliated institutions from Olympus, Ferring, and Abbvie. S.C.N. is a scientific co‐founder and shareholder of GenieBiome Ltd. S.C.N. receives patent royalties through her affiliated institutions. F.K.L.C., S.C.N., H.M.T. are named inventors of patent applications held by The CUHK and MagIC that cover the therapeutic and diagnostic use of microbiome but have no potential relevant financial or nonfinancial interests to disclose. The other authors have no conflicts of interest to declare.
Figures




References
-
- Rautava S, Luoto R, Salminen S, Isolauri E. Microbial contact during pregnancy, intestinal colonization and human disease. Nat Rev Gastroenterol Hepatol. 2012;9(12):565‐576. - PubMed
Associated data
LinkOut - more resources
Full Text Sources
