Phylogenomics of the rarest animals: a second species of Micrognathozoa identified by machine learning
- PMID: 39968621
- PMCID: PMC11836703
- DOI: 10.1098/rspb.2024.2867
Phylogenomics of the rarest animals: a second species of Micrognathozoa identified by machine learning
Abstract
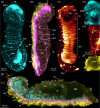
The latest animal phylum to be discovered, Micrognathozoa, constitutes a rare group of limnic meiofauna. These microscopic 'jaw animals' are among the smallest metazoans yet possess highly complex jaw structures. The single species of Micrognathozoa, Limnognathia maerski Kristensen and Funch, 2000, was first described from Greenland, later reported from a remote Subantarctic island and more recently discovered in the Pyrenees on the European continent. Successful collections of these three known populations facilitated investigations of the intraphylum relationships and species limits within Limnognathia for the first time. Through detailed anatomical comparisons, we substantiate the lack of morphological differences between the three geographically disjunct populations. With transcriptomic data from single specimens, we conducted the first intraphylum phylogenetic analyses and extensively tested species hypotheses using standard approaches and novel machine learning methods. Analyses clearly delimited the Subantarctic population, here described as Limnognathia desmeti sp. nov., the second species of Micrognathozoa, but did not definitively split the Greenland and Pyrenees populations as separate species. Divergence dating analysis suggests the disjunct distribution of Micrognathozoa is not human mediated but the result of long-distance dispersal raising questions about their dispersal capabilities and potential undiscovered populations.
Keywords: Spiralia; confocal laser scanning microscopy; integrated taxonomy; meiobenthos; phylotranscriptomics; species delimitation.
Conflict of interest statement
We declare we have no competing interests.
Figures


References
-
- De Smet WH. 2002. A new record of Limnognathia maerski Kristensen & Funch, 2000 (Micrognathozoa) from the subantarctic Crozet Islands, with redescription of the trophi. J. Zool. 258, 381–393. (10.1017/s095283690200153x) - DOI
-
- Sørensen MV. 2000. An SEM study of the jaws of Haplognathia rosea and Rastrognathia macrostoma (Gnathostomulida), with a preliminary comparison with the rotiferan trophi. Acta Zool. 81, 200. (10.1046/j.1463-6395.2000.00032.x) - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

