Exploring phenotypic and genotypic diversity among methicillin-resistant, vancomycin-resistant, and sensitive Staphylococcus aureus
- PMID: 39969287
- PMCID: PMC11688052
- DOI: 10.1097/MD.0000000000041051
Exploring phenotypic and genotypic diversity among methicillin-resistant, vancomycin-resistant, and sensitive Staphylococcus aureus
Abstract
Background: Methicillin-Resistant Staphylococcus aureus (MRSA) is a global concern owing to the increasing prevalence of multidrug-resistant (MDR) strains. Vancomycin has been the primary treatment for MRSA; however, Vancomycin-resistant strains are being increasingly reported worldwide. Therefore, comparative studies are essential to support antimicrobial stewardship and improving clinical management. Ultimately, the findings from this study are expected to inform treatment strategies and guide public health interventions effectively.
Material and methods: This study investigated the prevalence, antimicrobial resistance, and virulence characteristics of Vancomycin-sensitive S. aureus (VSSA) and Vancomycin-resistant S. aureus (VRSA) within MRSA strains. By employing a combination of phenotypic methods, such as antimicrobial susceptibility testing, and genotypic techniques, including molecular typing and identification of virulence genes, we obtained comprehensive insights into VRSA and VSSA profiles.
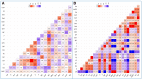
Results: Of 250 clinical samples, 62 (24.8%) were S. aureus and 27 (43.5%) were identified as MRSA. All MRSA isolates exhibited MDR patterns. Most MRSA strains were VSSA (20/27, 74.1%), while 7 (25.9%) were VRSA. The VRSA isolates showed more antimicrobial resistance than VSSA isolates; however, the VRSA isolates had less virulence than VSSA isolates. Linezolid was the most effective treatment, with a 3.7% resistance rate. A higher percentage of biofilm-producing MRSA (96.3%) was confirmed by both phenotypic and genotypic methods. All isolates, except one VRSA, showed multi-virulence patterns (harbored more than 3 virulence genes). High diversity and low clonality (D-value = 0.99) were found in both VSSA and VRSA. Based on our correlation findings, the emergence of vancomycin resistance could modify the association between antimicrobial resistance and virulence, potentially affecting the pathogenic profile of these strains. The study also revealed complex interactions among host factors (including age and gender), sample origin, antimicrobial resistance, biofilm production, and virulence genes.
Conclusion: This study highlights the alarming spread of MRSA and VRSA, which show significant resistance and virulence.
Copyright © 2024 the Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures





References
-
- Luzzago C, Locatelli C, Franco A, et al. . Clonal diversity virulence-associated genes and antimicrobial resistance profile of Staphylococcus aureus isolates from nasal cavities and soft tissue infections in wild ruminants in Italian Alps. Vet Microbiol. 2014;170:157–61. - PubMed
-
- Saleem N, Nawaz M, Ghafoor A, et al. . Phenotypic and molecular analysis of antibiotic resistance in lactobacilli of poultry origin from Lahore Pakistan. Pak Vet J. 2018;38:409.
-
- Shariati A, Dadashi M, Moghadam MT, van Belkum A, Yaslianifard S, Darban-Sarokhalil D. Global prevalence and distribution of vancomycin resistant vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Sci Rep. 2020;10:12689. - PMC - PubMed
-
- El-Aziz NKA, El-Hamid MIA, Bendary MM, El-Azazy AA, Ammar AM. Existence of vancomycin resistance among methicillin-resistant Staphylococcus aureus recovered from animal and human sources in Egypt. Slovenian Vet Res. 2018;55:221–30.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

