Bayesian classification of OXPHOS deficient skeletal myofibres
- PMID: 39970187
- PMCID: PMC11838899
- DOI: 10.1371/journal.pcbi.1012770
Bayesian classification of OXPHOS deficient skeletal myofibres
Abstract
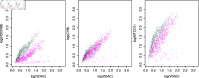
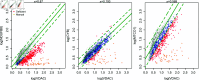
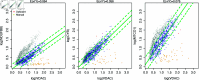
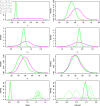
Mitochondria are organelles in most human cells which release the energy required for cells to function. Oxidative phosphorylation (OXPHOS) is a key biochemical process within mitochondria required for energy production and requires a range of proteins and protein complexes. Mitochondria contain multiple copies of their own genome (mtDNA), which codes for some of the proteins and ribonucleic acids required for mitochondrial function and assembly. Pathology arises from genetic defects in mtDNA and can reduce cellular abundance of OXPHOS proteins, affecting mitochondrial function. Due to the continuous turn-over of mtDNA, pathology is random and neighbouring cells can possess different OXPHOS protein abundance. Estimating the proportion of cells where OXPHOS protein abundance is too low to maintain normal function is critical to understanding disease severity and predicting disease progression. Currently, one method to classify single cells as being OXPHOS deficient is prevalent in the literature. The method compares a patient's OXPHOS protein abundance to that of a small number of healthy control subjects. If the patient's cell displays an abundance which differs from the abundance of the controls then it is deemed deficient. However, due to the natural variation between subjects and the low number of control subjects typically available, this method is inflexible and often results in a large proportion of patient cells being misclassified. These misclassifications have significant consequences for the clinical interpretation of these data. We propose a single-cell classification method using a Bayesian hierarchical mixture model, which allows for inter-subject OXPHOS protein abundance variation. The model accurately classifies an example dataset of OXPHOS protein abundances in skeletal muscle fibres (myofibres). When comparing the proposed and existing model classifications to manual classifications performed by experts, the proposed model results in estimates of the proportion of deficient myofibres that are consistent with expert manual classifications.
Copyright: © 2025 Childs et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
A stagewise response to mitochondrial dysfunction in mitochondrial DNA maintenance disorders.Biochim Biophys Acta Mol Basis Dis. 2024 Jun;1870(5):167131. doi: 10.1016/j.bbadis.2024.167131. Epub 2024 Mar 21. Biochim Biophys Acta Mol Basis Dis. 2024. PMID: 38521420
-
Coordinating mitochondrial translation with assembly of the OXPHOS complexes.Hum Mol Genet. 2024 May 22;33(R1):R47-R52. doi: 10.1093/hmg/ddae025. Hum Mol Genet. 2024. PMID: 38779773 Free PMC article. Review.
-
Failed upregulation of TFAM protein and mitochondrial DNA in oxidatively deficient fibers of chronic obstructive pulmonary disease locomotor muscle.Skelet Muscle. 2016 Feb 18;6:10. doi: 10.1186/s13395-016-0083-9. eCollection 2016. Skelet Muscle. 2016. PMID: 26893822 Free PMC article.
-
Mechanisms of mitochondrial diseases.Ann Med. 2012 Feb;44(1):41-59. doi: 10.3109/07853890.2011.598547. Epub 2011 Aug 2. Ann Med. 2012. PMID: 21806499
-
Significance of Mitochondria DNA Mutations in Diseases.Adv Exp Med Biol. 2017;1038:219-230. doi: 10.1007/978-981-10-6674-0_15. Adv Exp Med Biol. 2017. PMID: 29178079 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

