Disabling leading and lagging strand histone transmission results in parental histones loss and reduced cell plasticity and viability
- PMID: 39970210
- PMCID: PMC11837984
- DOI: 10.1126/sciadv.adr1453
Disabling leading and lagging strand histone transmission results in parental histones loss and reduced cell plasticity and viability
Abstract
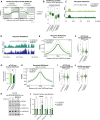
In the process of DNA replication, the first steps in restoring the chromatin landscape involve parental histone recycling and new histone deposition. Disrupting histone recycling to either the leading or lagging strand induces asymmetric histone inheritance, affecting epigenome maintenance and cellular identity. However, the order and kinetics of these effects remain elusive. Here, we use inducible mutants to dissect the early and late consequences of impaired histone recycling. Simultaneous disruption of both leading (POLE4) and lagging strand (MCM2-2A) recycling pathways impairs the transmission of parental histones to newly synthesized DNA, releasing some parental histones to the soluble pool. Subsequently, H3K27me3 accumulates aberrantly during chromatin restoration in a manner preceding gene expression changes. Loss of histone inheritance and the ensuing chromatin restoration defects alter gene expression in embryonic stem cells and challenge differentiation programs and cell viability. Our findings demonstrate the importance of efficient transmission of histone-based information during DNA replication for maintaining chromatin landscapes, differentiation potential, and cellular viability.
Figures



References
-
- Petryk N., Dalby M., Wenger A., Stromme C. B., Strandsby A., Andersson R., Groth A., MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361, 1389–1392 (2018). - PubMed
-
- Stewart-Morgan K. R., Petryk N., Groth A., Chromatin replication and epigenetic cell memory. Nat. Cell Biol. 22, 361–371 (2020). - PubMed
-
- Reverón-Gómez N., González-Aguilera C., Stewart-Morgan K. R., Petryk N., Flury V., Graziano S., Johansen J. V., Jakobsen J. S., Alabert C., Groth A., Accurate recycling of parental histones reproduces the histone modification landscape during DNA replication. Mol. Cell 72, 239–249.e5 (2018). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

