Biophysical and structural mechanisms of epilepsy-associated mutations in the S4-S5 Linker of KCNQ2 channels
- PMID: 39971736
- PMCID: PMC11845087
- DOI: 10.1080/19336950.2025.2464735
Biophysical and structural mechanisms of epilepsy-associated mutations in the S4-S5 Linker of KCNQ2 channels
Abstract
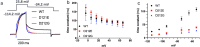
Mutations in KCNQ2 are linked to various neurological disorders, including neonatal-onset epilepsy. The severity of these conditions often correlates with the mutation's location and the biochemical properties of the altered amino acid side chains. Two mutations affecting aspartate at position 212 (D212) in the S4-S5 linker of KCNQ2 have been identified. Interestingly, while the charge-conserved D212E mutation leads to severe neonatal-onset developmental and epileptic encephalopathy (DEE), the more dramatic substitution to glycine (D212G) results in self-limited familial neonatal epilepsy (SLFNE), a much milder pathology. To elucidate the underlying mechanisms, we performed electrophysiological studies and in silico simulations to investigate these mutations' biophysical and structural effects. Our findings reveal that the D212E mutation stabilizes the channel in the voltage sensor down-state and destabilizes the up-state, leading to a rightward shift in the voltage-dependent activation curve, slower activation kinetics, and accelerated deactivation kinetics. This disruption in KCNQ2 voltage sensitivity persists even in the more physiologically relevant KCNQ2/3 heterotetrameric channels. In contrast, the D212G mutation primarily destabilizes the up-state, but its impact on voltage sensitivity is significantly reduced in KCNQ2/3 heterotetrameric channels. These findings provide key insights into the biophysical and structural basis of KCNQ2 D212 mutations and their contribution to epilepsy-related symptoms, offering a clearer understanding of how these mutations drive the varied clinical outcomes observed in patients.
Keywords: KCNQ; channelopathy; developmental and epileptic encephalopathy; gating; self-limited familial neonatal epilepsy.
Conflict of interest statement
Dr. Yen-Yu Yang is an Application Scientist at Thermo Fisher Scientific. No potential conflicts of interest were reported by any of the authors.
Figures




Similar articles
-
Distinctive mechanisms of epilepsy-causing mutants discovered by measuring S4 movement in KCNQ2 channels.Elife. 2022 Jun 1;11:e77030. doi: 10.7554/eLife.77030. Elife. 2022. PMID: 35642783 Free PMC article.
-
Dynamic PIP2 interactions with voltage sensor elements contribute to KCNQ2 channel gating.Proc Natl Acad Sci U S A. 2013 Dec 10;110(50):20093-8. doi: 10.1073/pnas.1312483110. Epub 2013 Nov 25. Proc Natl Acad Sci U S A. 2013. PMID: 24277843 Free PMC article.
-
Sequence determinants of subtype-specific actions of KCNQ channel openers.J Physiol. 2017 Feb 1;595(3):663-676. doi: 10.1113/JP272762. Epub 2016 Sep 23. J Physiol. 2017. PMID: 27506413 Free PMC article.
-
Flexible Stoichiometry: Implications for KCNQ2- and KCNQ3-Associated Neurodevelopmental Disorders.Dev Neurosci. 2021;43(3-4):191-200. doi: 10.1159/000515495. Epub 2021 Apr 1. Dev Neurosci. 2021. PMID: 33794528 Free PMC article. Review.
-
KCNQ2-Related Epilepsy: Genotype-Phenotype Relationship with Tailored Antiseizure Medication (ASM)-A Systematic Review.Neuropediatrics. 2023 Oct;54(5):297-307. doi: 10.1055/a-2060-4576. Epub 2023 Mar 22. Neuropediatrics. 2023. PMID: 36948217
References
-
- Miceli F, Soldovieri MV, Weckhuysen S, et al. KCNQ2-related disorders. In: Adam M, Everman D, Mirzaa G, Pagon R, Wallace SBean L, eds. GeneReviews(®). Seattle (WA): University of Washington. https://www.ncbi.nlm.nih.gov/books/NBK32534/. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
