Inhibition of Atg7 in intestinal epithelial cells drives resistance against Citrobacter rodentium
- PMID: 39971913
- PMCID: PMC11840101
- DOI: 10.1038/s41419-025-07422-5
Inhibition of Atg7 in intestinal epithelial cells drives resistance against Citrobacter rodentium
Abstract
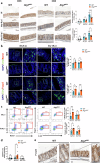
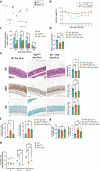
Autophagy, a cytoprotective mechanism in intestinal epithelial cells, plays a crucial role in maintaining intestinal homeostasis. Beyond its cell-autonomous effects, the significance of autophagy in these cells is increasingly acknowledged in the dynamic interplay between the microbiota and the immune response. In the context of colon cancer, intestinal epithelium disruption of autophagy has been identified as a critical factor influencing tumor development. This disruption modulates the composition of the gut microbiota, eliciting an anti-tumoral immune response. Here, we report that Atg7 deficiency in intestinal epithelial cells shapes the intestinal microbiota leading to an associated limitation of colitis induced by Citrobacter rodentium infection. Mice with an inducible, intestinal epithelial-cell-specific deletion of the autophagy gene, Atg7, exhibited enhanced clearance of C. rodentium, mitigated hyperplasia, and reduced pathogen-induced goblet cell loss. This protective effect is linked to a higher proportion of neutrophils and phagocytic cells in the early phase of infection. At later stages, it is associated with the downregulation of pro-inflammatory pathways and an increase in Th17 and Treg responses-immune responses known for their protective roles against C. rodentium infection, modulated by specific gut microbiota. Fecal microbiota transplantation and antibiotic treatment approaches revealed that the Atg7-deficiency-shapped microbiota, especially Gram-positive bacteria, playing a central role in driving resistance to C. rodentium infection. In summary, our findings highlight that inhibiting autophagy in intestinal epithelial cells contributes to maintaining homeostasis and preventing detrimental intestinal inflammation through microbiota-mediated colonization resistance against C. rodentium. This underscores the central role played by autophagy in shaping the microbiota in promoting immune-mediated resistance against enteropathogens.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–35. - PubMed
-
- Levy J, Cacheux W, Bara MA, L’Hermitte A, Lepage P, Fraudeau M, et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat Cell Biol. 2015;17:1062–73. - PubMed
-
- Roberti MP, Yonekura S, Duong CPM, Picard M, Ferrere G, Tidjani Alou M, et al. Chemotherapy-induced ileal crypt apoptosis and the ileal microbiome shape immunosurveillance and prognosis of proximal colon cancer. Nat Med. 2020;26:919–31. - PubMed
-
- Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol. 2018;20:521–7. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

