Unravelling genotype-phenotype correlations in Stargardt disease using patient-derived retinal organoids
- PMID: 39971915
- PMCID: PMC11840025
- DOI: 10.1038/s41419-025-07420-7
Unravelling genotype-phenotype correlations in Stargardt disease using patient-derived retinal organoids
Abstract
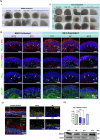
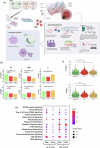
Stargardt disease is an inherited retinopathy affecting approximately 1:8000 individuals. It is characterised by biallelic variants in ABCA4 which encodes a vital protein for the recycling of retinaldehydes in the retina. Despite its prevalence and impact, there are currently no treatments available for this condition. Furthermore, 35% of STGD1 cases remain genetically unsolved. To investigate the cellular and molecular characteristics associated with STGD1, we generated iPSCs from two monoallelic unresolved (PT1 & PT2), late-onset STGD1 cases with the heterozygous complex allele - c.[5461-10 T > C;5603 A > T]. Both patient iPSCs and those from a biallelic affected control (AC) carrying -c.4892 T > C and c.4539+2001G > A, were differentiated to retinal organoids, which developed all key retinal neurons and photoreceptors with outer segments positive for ABCA4 expression. We observed patient-specific disruption to lamination with OPN1MW/LW+ cone photoreceptor retention in the retinal organoid centre during differentiation. Photoreceptor retention was more severe in the AC case affecting both cones and rods, suggesting a genotype/phenotype correlation. scRNA-Seq suggests retention may be due to the induction of stress-related pathways in photoreceptors. Whole genome sequencing successfully identified the missing alleles in both cases; PT1 reported c.-5603A > T in homozygous state and PT2 uncovered a rare hypomorph - c.-4685T > C. Furthermore, retinal organoids were able to recapitulate the retina-specific splicing defect in PT1 as shown by long-read RNA-seq data. Collectively, these results highlight the suitability of retinal organoids in STGD1 modelling. Their ability to display genotype-phenotype correlations enhances their utility as a platform for therapeutic development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests Ethics approval and consent to participate: All participants in this research study were identified and enlisted with informed consent by collaborators in the Marie-Skłodowska Curie Innovative Training Network – StarT (Grant no: 813490). All samples were collected in accordance with the tenets of the Declaration of Helsinki and written informed consent was obtained for all patients participating in the study.
Figures




References
-
- Blacharski. Fundus flavimaculatus. Retinal dystrophies and degenerations. 1988:135–59.
-
- Rotenstreich Y, Fishman GA, Anderson RJ. Visual acuity loss and clinical observations in a large series of patients with Stargardt disease. Ophthalmology. 2003;110:1151–8. - PubMed
-
- Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, Chidambaram A, et al. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997;15:236–46. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

