A novel identified epithelial ligand-receptor-associated gene signature highlights POPDC3 as a potential therapy target for non-small cell lung cancer
- PMID: 39971925
- PMCID: PMC11840029
- DOI: 10.1038/s41419-025-07410-9
A novel identified epithelial ligand-receptor-associated gene signature highlights POPDC3 as a potential therapy target for non-small cell lung cancer
Abstract
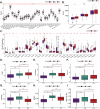
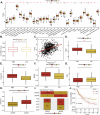
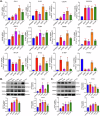
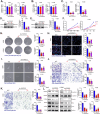
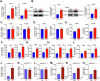
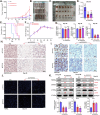
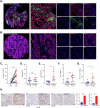
The tumor microenvironment (TME) is pivotal in non-small cell lung cancer (NSCLC) progression, influencing drug resistance and immune cell behavior through complex ligand-receptor (LR) interactions. This study developed an epithelial LR-related prognostic risk score (LRrisk) to identify biomarkers and targets in NSCLC. We identified twenty epithelial LRs with significant prognostic implications and delineated three molecular NSCLC subtypes with distinct outcomes, pathological characteristics, biological pathways, and immune profiles. The LRrisk model was constructed using twelve differentially expressed ligand-receptor interaction-related genes (LRGs), with a focus on POPDC3 (popeye domain-containing protein 3), which was overexpressed in NSCLC cells. Functional assays revealed that POPDC3 knockdown reduced cell proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT), while its overexpression promoted cancerous activities. In vivo, POPDC3 silencing hindered, and its overexpression accelerated the growth of NSCLC xenografts in nude mice. Additionally, high expression levels of POPDC3 in NSCLC tissues were associated with enhanced CD4+ T cell infiltration and increased PD-1 expression within the TME. Moreover, ectopic POPDC3 overexpression in C57BL/6 J mouse Lewis lung carcinoma (LLC) xenografts enhanced CD4+ T cell infiltration and PD-1 expression in the TME. This research establishes a robust epithelial LR-related signature, highlighting POPDC3 as a critical facilitator of NSCLC progression and a potential therapeutic target.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval: According to the Declaration of Helsinki, the Ethics Committee of Jiangsu University has approved this study.
Figures





References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. - PubMed
MeSH terms
Grants and funding
- 82072712/National Natural Science Foundation of China (National Science Foundation of China)
- 81773192/National Natural Science Foundation of China (National Science Foundation of China)
- 82304807/National Natural Science Foundation of China (National Science Foundation of China)
- 82273055/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical
Research Materials

