RNA neoantigen vaccines prime long-lived CD8+ T cells in pancreatic cancer
- PMID: 39972124
- PMCID: PMC11946889
- DOI: 10.1038/s41586-024-08508-4
RNA neoantigen vaccines prime long-lived CD8+ T cells in pancreatic cancer
Abstract
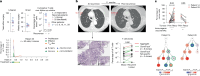
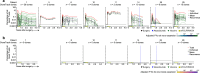
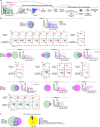
A fundamental challenge for cancer vaccines is to generate long-lived functional T cells that are specific for tumour antigens. Here we find that mRNA-lipoplex vaccines against somatic mutation-derived neoantigens may solve this challenge in pancreatic ductal adenocarcinoma (PDAC), a lethal cancer with few mutations. At an extended 3.2-year median follow-up from a phase 1 trial of surgery, atezolizumab (PD-L1 inhibitory antibody), autogene cevumeran1 (individualized neoantigen vaccine with backbone-optimized uridine mRNA-lipoplex nanoparticles) and modified (m) FOLFIRINOX (chemotherapy) in patients with PDAC, we find that responders with vaccine-induced T cells (n = 8) have prolonged recurrence-free survival (RFS; median not reached) compared with non-responders without vaccine-induced T cells (n = 8; median RFS 13.4 months; P = 0.007). In responders, autogene cevumeran induces CD8+ T cell clones with an average estimated lifespan of 7.7 years (range 1.5 to roughly 100 years), with approximately 20% of clones having latent multi-decade lifespans that may outlive hosts. Eighty-six percent of clones per patient persist at substantial frequencies approximately 3 years post-vaccination, including clones with high avidity to PDAC neoepitopes. Using PhenoTrack, a novel computational strategy to trace single T cell phenotypes, we uncover that vaccine-induced clones are undetectable in pre-vaccination tissues, and assume a cytotoxic, tissue-resident memory-like T cell state up to three years post-vaccination with preserved neoantigen-specific effector function. Two responders recurred and evidenced fewer vaccine-induced T cells. Furthermore, recurrent PDACs were pruned of vaccine-targeted cancer clones. Thus, in PDAC, autogene cevumeran induces de novo CD8+ T cells with multiyear longevity, substantial magnitude and durable effector functions that may delay PDAC recurrence. Adjuvant mRNA-lipoplex neoantigen vaccines may thus solve a pivotal obstacle for cancer vaccination.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: L.A.R., Z.M.S., B.D.G. and V.P.B. are inventors on patent applications related to work on antigen cross-reactivity and tracking vaccine-induced T cell clones. B.D.G. and V.P.B. are inventors on a patent application on neoantigen quality modelling. L.A.R. is an inventor of a patent related to oncolytic viral therapy. B.D.G. has received honoraria for speaking engagements from Merck, Bristol Meyers Squibb and Chugai Pharmaceuticals; has received research funding from Bristol Meyers Squibb, Merck and ROME Therapeutics; and has been a compensated consultant for Darwin Health, Merck, PMV Pharma, Shennon Biotechnologies, Synteny and Rome Therapeutics of which he is a co-founder. V.P.B. reports honoraria and research support from Genentech and research support from Bristol-Myers Squibb. A.S.E received royalties from Up-To-Date. A.V. reports research funding from Lilly, Verastem, BioMed Valley Discoveries, Bristol-Myers Squibb and Silenseed. A.C.W. reports the following: Histosonics, consulting and Ipsen, clinical trial funding. E.M.O. reports research funding to the institution from: Genentech/Roche, BioNTech, AstraZeneca, Arcus, Elicio, Parker Institute, NIH/NCI, Digestive Care and Break Through Cancer; consulting via Data and Safety Monitoring Board (DSMB) for: Arcus, Alligator, Agenus, BioNTech, Ipsen, Merck, Moma Therapeutics, Novartis, Syros, Leap Therapeutics, Astellas, BMS, Fibrogen, Revolution Medicine, Merus Agios (spouse), Genentech-Roche (spouse), Eisai (spouse) and Servier (Spouse). J.D. owns stock in Alnylam Pharmaceuticals, Arrowroot Acquisition and Ionis Pharmaceuticals. T.M. is a co-founder and holds equity in IMVAQ Therapeutics; is a consultant for Immunos Therapeutics, ImmunoGenesis and Pfizer; has research support from Bristol-Myers Squibb, Surface Oncology, Kyn Therapeutics, Infinity Pharmaceuticals, Peregrine Pharmaceuticals, Adaptive Biotechnologies, Leap Therapeutics and Aprea; and has patents on applications related to work on oncolytic viral therapy, alphavirus-based vaccine, neoantigen modelling, CD40, GITR, OX40, PD-1 and CTLA-4. J.D.W. is a consultant for Apricity, CellCarta, Ascentage Pharma, AstraZeneca, Bicara Therapeutics, Boehringer Ingelheim, Bristol-Myers Squibb, Daiichi Sankyo, Dragonfly, Georgiamune, Imvaq, Larkspur, Psioxus, Recepta, Tizona and Sellas. J.D.W. receives grant and research support from Bristol-Myers Squibb and Sephora. J.D.W. has equity in Apricity, Arsenal IO, Ascentage, Imvaq, Linneaus, Georgiamune, Maverick and Tizona Therapeutics. W.P. reports research funding to institution from: Merck, Astellas, Miracogen and Amgen; consultancy or advisory board activity for: Astellas and EXACT Therapeutics; honoraria for Continuing Medical Education (CME) from: American Physician Institute and Integrity. O.T. and U.S. are co-founders, management board members and employees at BioNTech. E.D., L.M. and F.M. are employees at BioNTech. I.R., M.Y. and I.M. are employees at Genentech. The other authors declare no competing interests.
Figures







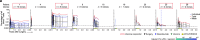




References
-
- Neoptolemos, J. P. et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. JAMA304, 1073–1081 (2010). - PubMed
-
- Uesaka, K. et al. Adjuvant chemotherapy of S-1 versus gemcitabine for resected pancreatic cancer: a phase 3, open-label, randomised, non-inferiority trial (JASPAC 01). Lancet388, 248–257 (2016). - PubMed
-
- Neoptolemos, J. P. et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, phase 3 trial. Lancet389, 1011–1024 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

