Dual regulation of mitochondrial fusion by Parkin-PINK1 and OMA1
- PMID: 39972141
- PMCID: PMC12399782
- DOI: 10.1038/s41586-025-08590-2
Dual regulation of mitochondrial fusion by Parkin-PINK1 and OMA1
Abstract
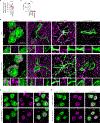
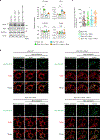
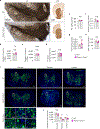
Mitochondrial stress pathways protect mitochondrial health from cellular insults1-8. However, their role under physiological conditions is largely unknown. Here, using 18 single, double and triple whole-body and tissue-specific knockout and mutant mice, along with systematic mitochondrial morphology analysis, untargeted metabolomics and RNA sequencing, we discovered that the synergy between two stress-responsive systems-the ubiquitin E3 ligase Parkin and the metalloprotease OMA1-safeguards mitochondrial structure and genome by mitochondrial fusion, mediated by the outer membrane GTPase MFN1 and the inner membrane GTPase OPA1. Whereas the individual loss of Parkin or OMA1 does not affect mitochondrial integrity, their combined loss results in small body size, low locomotor activity, premature death, mitochondrial abnormalities and innate immune responses. Thus, our data show that Parkin and OMA1 maintain a dual regulatory mechanism that controls mitochondrial fusion at the two membranes, even in the absence of extrinsic stress.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures












References
-
- Konig T et al. MIROs and DRP1 drive mitochondrial-derived vesicle biogenesis and promote quality control. Nat. Cell Biol 23, 1271–1286 (2021). - PubMed
-
- Vargas JNS, Hamasaki M, Kawabata T, Youle RJ & Yoshimori T The mechanisms and roles of selective autophagy in mammals. Nat. Rev. Mol. Cell Biol 24, 167–185 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

