Plasma proteome variation and its genetic determinants in children and adolescents
- PMID: 39972214
- PMCID: PMC11906355
- DOI: 10.1038/s41588-025-02089-2
Plasma proteome variation and its genetic determinants in children and adolescents
Abstract
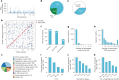
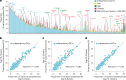
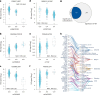
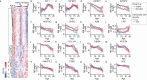
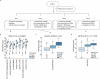
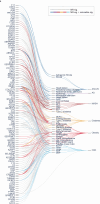
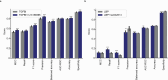
Our current understanding of the determinants of plasma proteome variation during pediatric development remains incomplete. Here, we show that genetic variants, age, sex and body mass index significantly influence this variation. Using a streamlined and highly quantitative mass spectrometry-based proteomics workflow, we analyzed plasma from 2,147 children and adolescents, identifying 1,216 proteins after quality control. Notably, the levels of 70% of these were associated with at least one of the aforementioned factors, with protein levels also being predictive. Quantitative trait loci (QTLs) regulated at least one-third of the proteins; between a few percent and up to 30-fold. Together with excellent replication in an additional 1,000 children and 558 adults, this reveals substantial genetic effects on plasma protein levels, persisting from childhood into adulthood. Through Mendelian randomization and colocalization analyses, we identified 41 causal genes for 33 cardiometabolic traits, emphasizing the value of protein QTLs in drug target identification and disease understanding.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.M. is an indirect investor in Evosep. L.N. and J.F. are employees of Novo Nordisk; however, this work was conducted while L.N. was a full-time employee at the University of Copenhagen. M.T. is a co-founder and board member of Evido. She is also a board member of the non-governmental organization Alcohol & Society. She receives speaker fees from Siemens Healthcare, Echosens, Norgine, Madrigal, Takeda and Tillotts Pharma as well as advisory fees from Boehringer Ingelheim, Astra Zeneca, Novo Nordisk and GSK. A.K. is a co-founder and board member of Evido. He has served as a speaker for Novo Nordisk, Norgine and Siemens, and has participated in advisory boards for Siemens, Boehringer Ingelheim and Novo Nordisk. Additionally, he receives research support from Astra Zeneca, Siemens, Nordic Bioscience and Echosense, all outside the submitted work. The other authors declare no competing interests.
Figures







References
MeSH terms
Substances
Grants and funding
- NNF15CC0001/Novo Nordisk Fonden (Novo Nordisk Foundation)
- NNF15OC0016692/Novo Nordisk Fonden (Novo Nordisk Foundation)
- NNF20OC0059393/Novo Nordisk Fonden (Novo Nordisk Foundation)
- NNF18SA0034956/Novo Nordisk Fonden (Novo Nordisk Foundation)
- NNF21SA0072102/Novo Nordisk Fonden (Novo Nordisk Foundation)
LinkOut - more resources
Full Text Sources

