Pro-inflammatory macrophages produce mitochondria-derived superoxide by reverse electron transport at complex I that regulates IL-1β release during NLRP3 inflammasome activation
- PMID: 39972217
- PMCID: PMC11946910
- DOI: 10.1038/s42255-025-01224-x
Pro-inflammatory macrophages produce mitochondria-derived superoxide by reverse electron transport at complex I that regulates IL-1β release during NLRP3 inflammasome activation
Abstract
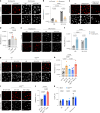
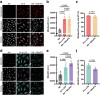
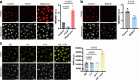
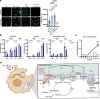
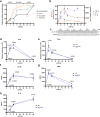
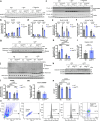
Macrophages stimulated by lipopolysaccharide (LPS) generate mitochondria-derived reactive oxygen species (mtROS) that act as antimicrobial agents and redox signals; however, the mechanism of LPS-induced mitochondrial superoxide generation is unknown. Here we show that LPS-stimulated bone-marrow-derived macrophages produce superoxide by reverse electron transport (RET) at complex I of the electron transport chain. Using chemical biology and genetic approaches, we demonstrate that superoxide production is driven by LPS-induced metabolic reprogramming, which increases the proton motive force (∆p), primarily as elevated mitochondrial membrane potential (Δψm) and maintains a reduced CoQ pool. The key metabolic changes are repurposing of ATP production from oxidative phosphorylation to glycolysis, which reduces reliance on F1FO-ATP synthase activity resulting in a higher ∆p, while oxidation of succinate sustains a reduced CoQ pool. Furthermore, the production of mtROS by RET regulates IL-1β release during NLRP3 inflammasome activation. Thus, we demonstrate that ROS generated by RET is an important mitochondria-derived signal that regulates macrophage cytokine production.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures








References
-
- Kravchuk, V. et al. A universal coupling mechanism of respiratory complex I. Nature10.1038/s41586-022-05199-7 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

