EGFR-to-Src family tyrosine kinase switching in proliferating-DTP TNBC cells creates a hyperphosphorylation-dependent vulnerability to EGFR TKI
- PMID: 39972345
- PMCID: PMC11841279
- DOI: 10.1186/s12935-025-03691-4
EGFR-to-Src family tyrosine kinase switching in proliferating-DTP TNBC cells creates a hyperphosphorylation-dependent vulnerability to EGFR TKI
Abstract
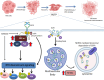
Triple-Negative Breast Cancer (TNBC) is the most aggressive type of breast malignancy, with chemotherapy as the only mainstay treatment. TNBC patients have the worst prognoses as a large fraction of them do not achieve complete pathological response post-treatment and develop drug-resistant residual disease. Molecular mechanisms that trigger proliferation in drug-resistant chemo-residual TNBC cells are poorly understood due to the lack of investigations using clinically relevant cellular models. In this study, we have established TNBC subtype-specific cellular models of proliferating drug-tolerant persister (PDTP) cells using different classes of chemotherapeutic agents that recapitulate clinical residual disease with molecular heterogeneity. Analysis of total phospho-tyrosine signals in TNBC PDTPs showed an enhanced phospho-tyrosine content compared to the parental cells (PC). Interestingly, using mass-spectrometry analysis, we identified a dramatic decrease in epidermal growth factor receptor (EGFR) expression in the PDTPs, while the presence of hyper-activated tyrosine phosphorylation of EGFR compared to PC. Further, we show that EGFR has enhanced lysosomal trafficking in PDTPs with a concomitant increase in N-Myc Downstream Regulated-1 (NDRG1) expression that co-localizes with EGFR to mediate receptor degradation. More surprisingly, we found that reduced protein levels of EGFR are coupled with a robust increase in Src family kinases, including Lyn and Fyn kinases, that creates a hyper-phosphorylation state of EGFR-Src tyrosine kinases axis in PDTPs and mediates downstream over-activation of STAT3, AKT and MAP kinases. Moreover, paclitaxel-derived PDTPs show increased sensitivity to EGFR TKI Gefitinib and its combination with paclitaxel selectively induced cell death in Paclitaxel-derived PDTP (PDTP-P) TNBC cells and 3D spheroids by strongly downregulating phosphorylation of EGFR-Src with concomitant downregulation of Lyn and Fyn tyrosine kinases. Collectively, this study identifies a unique hyper-phosphorylation cellular state of TNBC PDTPs established by switching of EGFR-Src family tyrosine kinases, creating a vulnerability to EGFR TKI.
Keywords: Combination therapy; EGFR; Proliferating drug-tolerant persister (PDTPs); Src family kinases; TNBC.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures






References
-
- Penault-Llorca F, Viale G. Pathological and molecular diagnosis of triple-negative breast cancer: a clinical perspective. Ann Oncol. 2012;23:vi19–22. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

