Cullin-RING ligase BioE3 reveals molecular-glue-induced neosubstrates and rewiring of the endogenous Cereblon ubiquitome
- PMID: 39972349
- PMCID: PMC11841277
- DOI: 10.1186/s12964-025-02091-5
Cullin-RING ligase BioE3 reveals molecular-glue-induced neosubstrates and rewiring of the endogenous Cereblon ubiquitome
Abstract
Background: The specificity of the ubiquitination process is mediated by the E3 ligases. Discriminating genuine substrates of E3s from mere interacting proteins is one of the major challenges in the field. We previously developed BioE3, a biotin-based approach that uses BirA-E3 fusions together with ubiquitin fused to a low-affinity AviTag to obtain a site-specific and proximity-dependent biotinylation of the substrates. We proved the suitability of BioE3 to identify targets of RING and HECT-type E3 ligases.
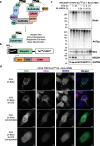
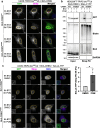
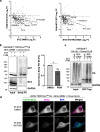
Methods: BioE3 experiments were performed in HEK293FT and U2OS stable cell lines expressing TRIPZ-bioGEFUb transiently transfected with BirA-cereblon (CRBN). Cells were seeded using biotin-free media, followed later by a short-biotin pulse. We evaluated the applicability of the BioE3 system to CRBN and molecular glues by Western blot and confocal microscopy, blocking the proteasome with bortezomib, inhibiting NEDDylation with MLN4924 and treating the cells with pomalidomide. For the identification of endogenous substrates and neosubstrates we analyzed the eluates of streptavidin pull-downs of BioE3 experiments by LC-MS/MS. Analysis of targets for which ubiquitination changes significantly upon treatment was done using two-sided Student's t-test. Orthogonal validations were performed by histidine pull-down, GFP-trap and computational modelling.
Results: Here we demonstrate that BioE3 is suitable for the multi-protein complex Cullin-RING E3s ligases (CRLs), the most utilized E3-type for targeted protein degradation (TPD) strategies. Using CRBN as proof of concept, one of the substrate receptors of CRL4 E3 ligase, we identified both endogenous substrates and novel neosubstrates upon pomalidomide treatment, including CSDE1 which contains a G-loop motif potentially involved in the binding to CRBN in presence of pomalidomide. Importantly, we observed a major rearrangement of the endogenous ubiquitination landscape upon treatment with this molecular glue.
Conclusions: The ability of BioE3 to detect and compare both substrates and neosubstrates, as well as how substrates change in response to treatments, will facilitate both on-target and off-target identifications and offer a broader characterization and validation of TPD compounds, like molecular glues and PROTACs.
Keywords: E3 ligases; Immunomodulatory drugs; Molecular Glue; Targeted Protein Degradation; Ubiquitin.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: C.G. is co-founder and Drug Discovery Scientific Advisor of Oniria Therapeutics. The other authors declare that they have no competing interests.
Figures


References
-
- Lange SM, Armstrong LA, Kulathu Y. Deubiquitinases: From mechanisms to their inhibition by small molecules. Mol Cell. 2022;82(1):15–29. - PubMed
-
- Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. 2017;86:129–57. - PubMed
-
- Morreale FE, Walden H. Types of ubiquitin ligases. Cell. 2016;165(1):248–e1. - PubMed
MeSH terms
Substances
Grants and funding
- FPU20/05282/Ministerio de Universidades
- 765445-EU, UbiCODE Program/HORIZON EUROPE Marie Sklodowska-Curie Actions
- PRE2022-104553/Ministerio de Ciencia e Innovación
- PRE2018-086230/Ministerio de Ciencia e Innovación
- PRE2021-099359/Ministerio de Ciencia e Innovación
- PREP2023-000294/Ministerio de Ciencia e Innovación
- ProteoCure CA20113/Cooperation for Science & Technology COST Action
- 2021SGR00671/Computational Biology Drug Design Consolidated Research Group, Generalitat de Catalunya
- CNS2022-135307/Ministerio de Ciencia, Innovación y Universidades
- PID2020-117333GB-I00/Ministerio de Ciencia, Innovación y Universidades
- PID2023-147399NB-I00, PID2020-114178GB-I00, CEX2021-001136-S, CEX2021-001202-M/Ministerio de Ciencia, Innovación y Universidades
- BCV-2021-3-0006, BCV-2021-2-0005/Red Española de Supercomputación
- 6/12/TT/2023 /00001/Diputación Foral de Bizkaia
LinkOut - more resources
Full Text Sources

