Spatial analysis of a complete DIPG-infiltrated brainstem reveals novel ligand-receptor mediators of tumour-to-TME crosstalk
- PMID: 39972389
- PMCID: PMC11837654
- DOI: 10.1186/s40478-025-01952-x
Spatial analysis of a complete DIPG-infiltrated brainstem reveals novel ligand-receptor mediators of tumour-to-TME crosstalk
Abstract
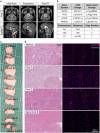
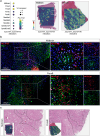
Previous studies have highlighted the capacity of brain cancer cells to functionally interact with the tumour microenvironment (TME). This TME-cancer crosstalk crucially contributes to tumour cell invasion and disease progression. In this study, we performed spatial transcriptomic sequencing analysis of a complete annotated tumour-infiltrated brainstem from a single diffuse intrinsic pontine glioma (DIPG) patient. Gene signatures from ten sequential tumour regions were analysed to assess mechanisms of disease progression and oncogenic interactions with the TME. We identified four distinct tumour subpopulations and assessed respective ligand-receptor pairs that actively promote DIPG tumour progression via crosstalk with endothelial, neuronal and immune cell communities. Our analysis found potential targetable mediators of tumour-to-TME communication, including members of the complement component system and the neuropeptide/GPCR ligand-receptor pair ADCYAP1-ADCYAP1R1. These interactions could influence DIPG tumour progression and represent novel therapeutic targets.
Keywords: ADCYAP1-ADCYAP1R1; DIPG; Ligand-receptor pairs; Spatial transcriptomic sequencing; Tumour-CNS crosstalk.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: This study was performed in line with ethical approval from the QIMR human research ethics (HREC) committee under project P3420. Consent to publish: Written informed consent was obtained from the patient’s legal representative. The authors affirm that human research participants provided informed consent for publication of the images in Figure(s) 1a and 1c. Competing interests: The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

