CircRNA hsa_circ_0004781 promoted cell proliferation by acting as a sponge for miR-9-5p and miR-338-3p and upregulating KLF5 and ADAM17 expression in pancreatic ductal adenocarcinoma
- PMID: 39972489
- PMCID: PMC11841339
- DOI: 10.1186/s12935-025-03687-0
CircRNA hsa_circ_0004781 promoted cell proliferation by acting as a sponge for miR-9-5p and miR-338-3p and upregulating KLF5 and ADAM17 expression in pancreatic ductal adenocarcinoma
Abstract
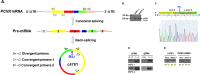
Background: Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive types of solid tumor, and novel strategies must be developed for treating it. Previous studies predominantly utilized circular RNA (circRNA) expression plasmids incorporating Alu elements to facilitate the indirect expression of circRNA.
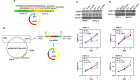
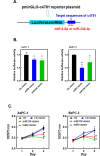
Methods: Public databases and bioinformatics tools were used to identify hsa_circ_0004781 that is highly expressed in PDAC and its potential microRNA (miRNA) targets and corresponding mRNA targets. Real hsa_circ_0004781, which is identical to the native form of hsa_circ_0004781 without any exogenous sequences, was prepared through in vitro transcription by using a ribozyme and ion-pair reversed-phase high-performance liquid chromatography (IP-RP HPLC). The biological functions of hsa_circ_0004781 were evaluated using loss-of-function and gain-of-function approaches with circRNA expression plasmids and real hsa_circ_0004781.
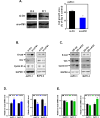
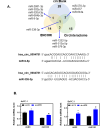
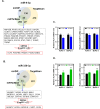
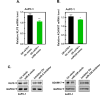
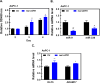
Results: Knockdown of hsa_circ_0004781 inhibited the proliferation and migration of PDAC cells, whereas its overexpression produced opposite effects. Hsa_circ_0004781 was identified as a sponge for miR-9-5p and miR-338-3p, and its expression was negatively correlated with that of these miRNAs. Among the targets of miR-9-5p and miR-338-3p, Kruppel-like factor 5 (KLF5) and a disintegrin and metalloproteinase domain 17 (ADAM17) were negatively correlated with survival in patients with PDAC and were inversely regulated by these miRNAs. Furthermore, real hsa_circ_0004781 exhibited the same effects as those of the circRNA expression plasmids.
Conclusions: This study is the first to use real circRNAs to validate results obtained using circRNA expression plasmids. The results suggest that hsa_circ_0004781 functions as an oncogene, promoting the proliferation of PDAC cells through the miR-9-5p/KLF5 and miR-338-3p/ADAM17 axes. Therefore, hsa_circ_0004781 might be a therapeutic target for PDAC.
Keywords: ADAM17; Hsa_circ_0004781; KLF5; Pancreatic ductal adenocarcinoma; Real circRNA; miR-338-3p; miR-9-5p.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Circular RNA hsa_circ_0007367 promotes the progression of pancreatic ductal adenocarcinoma by sponging miR-6820-3p and upregulating YAP1 expression.Cell Death Dis. 2022 Aug 25;13(8):736. doi: 10.1038/s41419-022-05188-8. Cell Death Dis. 2022. PMID: 36008392 Free PMC article.
-
Reveal the potential molecular mechanism of circRNA regulating immune-related mRNA through sponge miRNA in the occurrence and immune regulation of papillary thyroid cancer.Ann Med. 2023;55(2):2244515. doi: 10.1080/07853890.2023.2244515. Ann Med. 2023. PMID: 37603701 Free PMC article.
-
Whole-transcriptome analysis reveals a potential hsa_circ_0001955/hsa_circ_0000977-mediated miRNA-mRNA regulatory sub-network in colorectal cancer.Aging (Albany NY). 2020 Mar 28;12(6):5259-5279. doi: 10.18632/aging.102945. Epub 2020 Mar 28. Aging (Albany NY). 2020. PMID: 32221048 Free PMC article.
-
Silencing circular RNA hsa_circ_0000977 suppresses pancreatic ductal adenocarcinoma progression by stimulating miR-874-3p and inhibiting PLK1 expression.Cancer Lett. 2018 May 28;422:70-80. doi: 10.1016/j.canlet.2018.02.014. Epub 2018 Feb 15. Cancer Lett. 2018. Retraction in: Cancer Lett. 2018 Dec 1;438:232. doi: 10.1016/j.canlet.2018.09.027. PMID: 29454093 Retracted.
-
Upregulated circRNA hsa_circ_0071036 promotes tumourigenesis of pancreatic cancer by sponging miR-489 and predicts unfavorable characteristics and prognosis.Cell Cycle. 2021 Feb;20(4):369-382. doi: 10.1080/15384101.2021.1874684. Epub 2021 Jan 28. Cell Cycle. 2021. PMID: 33507122 Free PMC article.
References
-
- Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–91. - PubMed
-
- Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al. Oncogenic role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal translocations. Cell. 2016;165(2):289–302. - PubMed
-
- Borran S, Ahmadi G, Rezaei S, Anari MM, Modabberi M, Azarash Z, et al. Circular RNAs: New players in thyroid cancer. Pathol Res Pract. 2020;216(10):153217. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

