This is a preprint.
A novel lncRNA FAM151B-DT regulates autophagy and degradation of aggregation prone proteins
- PMID: 39974060
- PMCID: PMC11838976
- DOI: 10.1101/2025.01.22.25320997
A novel lncRNA FAM151B-DT regulates autophagy and degradation of aggregation prone proteins
Abstract
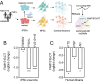
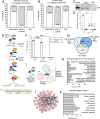
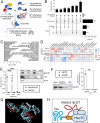
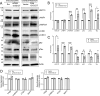
Neurodegenerative diseases share common features of protein aggregation along with other pleiotropic traits, including shifts in transcriptional patterns, neuroinflammation, disruptions in synaptic signaling, mitochondrial dysfunction, oxidative stress, and impaired clearance mechanisms like autophagy. However, key regulators of these pleotropic traits have yet to be identified. Here, we discovered a novel long non-coding RNA (lncRNA), FAM151B-DT, that is reduced in a stem cell model of frontotemporal dementia with tau inclusions (FTLD-tau) and in brains from FTLD-tau, progressive supranuclear palsy, Alzheimer's disease, and Parkinson's disease patients. We show that silencing FAM151B-DT in vitro is sufficient to enhance tau aggregation. To begin to understand the mechanism by which FAM151B-DT mediates tau aggregation and contributes to several neurodegenerative diseases, we deeply characterized this novel lncRNA and found that FAM151B-DT resides in the cytoplasm where it interacts with tau, α-synuclein, HSC70, and other proteins enriched in protein homeostasis. When silenced, FAM151B-DT blocks autophagy, leading to the accumulation of tau and α-synuclein. Importantly, we discovered that increasing FAM151B-DT expression is sufficient to promote autophagic flux, reduce phospho-tau and α-synuclein, and reduce tau aggregation. Overall, these findings pave the way for further exploration of FAM151B-DT as a promising molecular target for several neurodegenerative diseases.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
