The complete mitochondrial genome of a ground beetle, Chlaenius naeviger (Carabidae: Harpalinae: Chlaeniini), from South Korea
- PMID: 39974742
- PMCID: PMC11837945
- DOI: 10.1080/23802359.2025.2466587
The complete mitochondrial genome of a ground beetle, Chlaenius naeviger (Carabidae: Harpalinae: Chlaeniini), from South Korea
Abstract
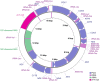
Chlaenius naeviger Morawitz, 1862 is a ground beetle species with a potential as a biological control agent for agricultural pests. In this study, we sequenced and annotated the complete mitochondrial genome (mitogenome) of C. naeviger, which is 16,594 bp in length and comprises 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and an A + T rich region. Furthermore, we used the nucleotide sequences of 13 PCGs to reconstruct a maximum likelihood phylogenetic tree of the family Carabidae, which revealed a close relationship between Chlaeniini and Panagaeini (BP 100). Our findings shed light on the phylogenetic relationships among the harpaline tribes.
Keywords: Chlaeniini; Chlaenius naeviger; ground beetle; mitochondrial genome; phylogeny.
© 2025 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Chaolong H, Yin T, Kanglai H, Zhenying W.. 2020. Predation of Chlaenius bioculatus larvae to larvae of Spodoptera frugiperda (Lepidoptera: noctuidae). Chinese J Biol Control. 36(4):507.
-
- Choudhuri JCB, Misra MP.. 1981. Chlaenius rayotus Bates (Coleoptera: Carabidae: Chlaeniini)-a new predator of Hyblaea puera Cramer (Lepidoptera: Hyblaeidae) from Walayar Reserve Forests. Kerala State. Indian Forester. 107(1):63–65.
LinkOut - more resources
Full Text Sources
Other Literature Sources
