This is a preprint.
The SP140-RESIST pathway regulates interferon mRNA stability and antiviral immunity
- PMID: 39974928
- PMCID: PMC11838211
- DOI: 10.1101/2024.08.28.610186
The SP140-RESIST pathway regulates interferon mRNA stability and antiviral immunity
Update in
-
SP140-RESIST pathway regulates interferon mRNA stability and antiviral immunity.Nature. 2025 Jul;643(8074):1372-1380. doi: 10.1038/s41586-025-09152-2. Epub 2025 Jun 11. Nature. 2025. PMID: 40500448 Free PMC article.
Abstract
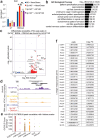
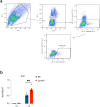
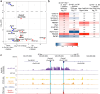
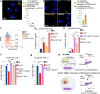
Type I interferons (IFN-Is) are essential for antiviral immunity but must be tightly regulated1-3. The conserved transcriptional repressor SP140 inhibits interferon beta (Ifnb1) expression via an unknown mechanism4,5. Here we report that SP140 does not directly repress Ifnb1 transcription. Instead, SP140 negatively regulates Ifnb1 mRNA stability by directly repressing the expression of a previously uncharacterized regulator we call RESIST (REgulated Stimulator of Interferon via Stabilization of Transcript, previously annotated as Annexin-2 Receptor). RESIST promotes Ifnb1 mRNA stability by counteracting Ifnb1 mRNA destabilization mediated by the Tristetraprolin (TTP) family of RNA-binding proteins and the CCR4-NOT deadenylase complex. SP140 localizes within nuclear bodies, punctate structures that play important roles in silencing DNA virus gene expression in the nucleus4. Consistent with this observation, we found that SP140 inhibits replication of the gammaherpesvirus MHV68. The antiviral activity of SP140 was independent of its ability to regulate Ifnb1. Our results establish dual antiviral and interferon regulatory functions for SP140. We propose that SP140 and RESIST participate in antiviral effector-triggered immunity6,7.
Conflict of interest statement
Competing Interests R.E.V. is on the Scientific Advisory Boards of Tempest Therapeutics and X-biotix.
Figures











References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
