This is a preprint.
C12ORF57: a novel principal regulator of synaptic AMPA currents and excitatory neuronal homeostasis
- PMID: 39974932
- PMCID: PMC11838199
- DOI: 10.1101/2025.01.08.632037
C12ORF57: a novel principal regulator of synaptic AMPA currents and excitatory neuronal homeostasis
Abstract
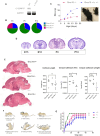
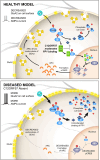
Objective: Excitatory neuronal homeostasis is crucial for neuronal survival, circuit function, and plasticity. Disruptions in this form of homeostasis are believed to underpin a variety of neuronal conditions including intellectual disability, epilepsy, and autism. However, the underlying genetic and molecular mechanisms maintaining this homeostasis remain poorly understood. Biallelic recurrent loss of function mutations in C12ORF57, an evolutionarily conserved X amino acid novel open reading frame, underlie Temtamy syndrome (TS)-a neurodevelopmental disorder characterized by epilepsy, dysgenesis of the corpus callosum, and severe intellectual disability.
Methods: Through multiple lines of inquiry, we establish that C12ORF57/GRCC10 plays an unexpected central role in synaptic homeostatic downscaling in response to elevated activity, uncovering a novel mechanism for neuronal excitatory homeostasis. To probe these mechanisms, we developed a new knockout (KO) mouse model of the gene's murine ortholog, Grcc10 as well as cellular and in vitro assays.
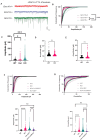
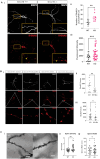
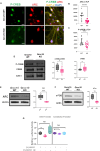
Results: Grcc10 KO mice exhibit the characteristic phenotypic features seen in human TS patients, including increased epileptiform activity. Corresponding with the enhanced seizure susceptibility, hippocampal neurons in these mice exhibited significantly increased AMPA receptor expression levels and higher amplitude of miniature excitatory postsynaptic currents (mEPSCs). We further found that GRCC10/C12ORF57 modulates the activity of calcium/calmodulin dependent kinase 4 (CAMK4) and thereby regulates the expression of CREB and ARC.
Interpretation: Our study suggests through this novel mechanism, deletion of Grcc10 disrupts the characteristic synaptic AMPA receptor downscaling that accompanies increased activity in glutamatergic neurons.
Keywords: AMPA receptors; ASD; C12ORF57; CAMK4; Temtamy syndrome; epilepsy.
Conflict of interest statement
Competing Interests The authors declare no competing interests.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
