This is a preprint.
Complex exchanges among plasmids and clonal expansion of lineages shape the population structure and virulence of Borrelia burgdorferi
- PMID: 39974970
- PMCID: PMC11838331
- DOI: 10.1101/2025.01.29.635312
Complex exchanges among plasmids and clonal expansion of lineages shape the population structure and virulence of Borrelia burgdorferi
Abstract
Background: In the United States, Borrelia burgdorferi (Bb) is the principal etiologic agent of Lyme disease. The complex structure of Bb genomes has posed challenges for genomic studies because homology among the bacterium's many plasmids, which account for ~40% of the genome by length, has made them difficult to sequence and assemble.
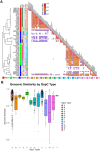
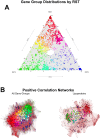
Results: We used long-read sequencing to generate near-complete assemblies of 62 isolates of human-derived Bb and collected public genomes with plasmid sequences. We characterized genetic diversity and population structure in the resulting set of 82 plasmid-complete Borrelia burgdorferi sensu stricto genomes. The Bb core genome is encoded by a chromosome and the conserved plasmids cp26, lp54, and lp17; the accessory genome is encoded by all other plasmids and the distal arm of the chromosome. Near-complete genomes reveal that the most granular Bb genotypes are clonal expansions of complex rearrangements among accessory genome elements. Ribosomal spacer types (RST) represent multiple collections of such genotypes, whereas OspC types are usually clonal. Structural rearrangements are non-randomly distributed throughout the genome, with cp32 plasmids undergoing dense exchanges and most linear plasmids, except lp54, sharing blocks among themselves and with the distal arm of the chromosome. OspC type A strains, known to possess greater virulence in humans, are distinguished by the presence of lp28-1 and lp56. Rearrangements among plasmids tended to preserve gene content, suggesting functional constraints among gene networks. Using k-partite graph decompositions, we identified gene sets with correlation patterns suggestive of conserved functional modules.
Conclusions: Long-read assemblies reveal that Bb population genetic structure results from clonal expansion of lineages that have undergone complex rearrangements among plasmid-encoded accessory genome elements. Genetic structure is preserved among genes even when plasmid rearrangements occur, suggesting that selection among epistatic loci maintains functional genetic networks. The analysis of near-complete genomes assembled using long-read sequencing methods advances our understanding of Bb biology and Lyme disease pathogenesis by providing the first detailed view of population variation in previously inaccessible areas of the Bb genome.
Keywords: Borrelia burgdorferi; Lyme disease; Pathogenicity; Virulence; Whole Genome Sequencing; genomics; lipoproteins; recombination.
Conflict of interest statement
P.C.S. is a co-founder of, shareholder in, and consultant to Sherlock Biosciences and Delve Bio, as well as a board member of and shareholder in Danaher Corporation. K.S. served as a consultant for T2 Biosystems, Roche, BioMerieux, and NYS Biodefense Fund, for the development of a diagnostic assay in Lyme borreliosis. F.S. served on the scientific advisory board for Roche on Lyme disease serological diagnostics and on the scientific advisory board for Pfizer on Lyme disease vaccine, and is an unpaid member of the steering committee of the ESCMID Study Group on Lyme Borreliosis/ESGBOR. J.A.B. has received research funding to his institution from Analog Devices Inc., Zeus Scientific, Immunetics, Pfizer, DiaSorin, bioMerieux and the Steven & Alexandra Cohen Foundation, and has been a paid consultant to T2 Biosystems, DiaSorin, Flightpath Biosciences and Roche Diagnostics. G.P.W. reports receiving research grants from Biopeptides, Corp. He has been an expert witness in malpractice cases involving Lyme disease and babesiosis; and is an unpaid board member of the non-profit American Lyme Disease Foundation. J.A.B. and J.E.L. are co-authors on a provisional patent application for the diagnosis of Lyme Disease unrelated to this work.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
