This is a preprint.
Clonal relationships between Tph and Tfh cells in patients with SLE and in murine lupus
- PMID: 39974998
- PMCID: PMC11838332
- DOI: 10.1101/2025.01.27.635189
Clonal relationships between Tph and Tfh cells in patients with SLE and in murine lupus
Abstract
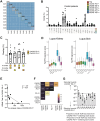
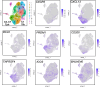
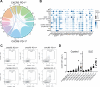
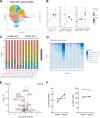
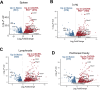
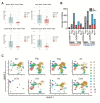
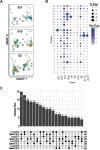
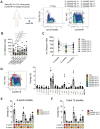
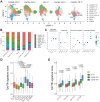
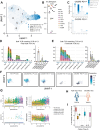
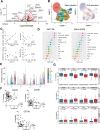
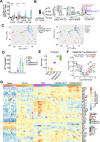
Pathologic T cell-B cell interactions drive disease in systemic lupus erythematosus (SLE). The T cells that activate B cell responses include T peripheral helper (Tph) and T follicular helper (Tfh) cells, yet the developmental and clonal relationships between these B cell-helper T cell populations are unclear. Here we use T cell receptor (TCR) profiling to demonstrate substantial clonal overlap between Tph and Tfh cells in the circulation of patients with SLE. Expanded Tph and Tfh cell clones persist over the course of 1 year in patients with a new diagnosis of SLE, and clones are observed to shift both from Tfh to Tph cells and from Tph to Tfh cells over time. High resolution analysis of cells sorted as Tph cells (CXCR5- PD-1hi) from SLE patients revealed considerable heterogeneity among these cells and highlighted a subpopulation of cells with transcriptomic features of activated B cell-helper T cells. This cell population, marked by expression of TOX and CXCL13, was found in both sorted Tph and Tfh cells, and was clonally linked in these two populations. Analysis of B cell-helper T cells in murine pristane-induced lupus demonstrated similar populations of Tph and Tfh cells in both lung and spleen with strong clonal overlap. T cell-specific loss of Bcl6 prevented accumulation of Tfh cells and reduced accumulation of Tph cells in pristane-treated mice, indicating a role for Bcl6 in the survival and expansion of both populations. Together, these observations demonstrate a shared developmental path among pathologically expanded Tph and Tfh cells in SLE. The persistence of expanded Tph and Tfh cells clones over time may explain the lack of stable tolerance induction by immunosuppressive medications or by B cell depletion.
Keywords: Systemic lupus erythematosus; T cell receptor repertoire; T follicular helper cell; T peripheral helper cell.
Conflict of interest statement
Competing interests The work was performed in part with grant support from Merck & Co., Inc. D.A.R. reports grant support from Janssen and Bristol-Myers Squibb outside of the current report, and reports personal fees from AstraZeneca, Pfizer, Merck, Abbvie, Simcere, Biogen, and Bristol-Myers Squibb. He is co-inventor on a patent using T peripheral helper cells as a biomarker of autoimmune diseases. Y.Q., M.A.S., S.E.A., and M.C.L. are employees of Merck & Co., Inc.
Figures




References
-
- Tsokos G. C., Systemic lupus erythematosus. N. Engl. J. Med. 365, 2110–2121 (2011). - PubMed
-
- Rao D. A., Gurish M. F., Marshall J. L., Slowikowski K., Fonseka C. Y., Liu Y., Donlin L. T., Henderson L. A., Wei K., Mizoguchi F., Teslovich N. C., Weinblatt M. E., Massarotti E. M., Coblyn J. S., Helfgott S. M., Lee Y. C., Todd D. J., Bykerk V. P., Goodman S. M., Pernis A. B., Ivashkiv L. B., Karlson E. W., Nigrovic P. A., Filer A., Buckley C. D., Lederer J. A., Raychaudhuri S., Brenner M. B., Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 542, 110–114 (2017). - PMC - PubMed
-
- Bocharnikov A. V., Keegan J., Wacleche V. S., Cao Y., Fonseka C. Y., Wang G., Muise E. S., Zhang K. X., Arazi A., Keras G., Li Z. J., Qu Y., Gurish M. F., Petri M., Buyon J. P., Putterman C., Wofsy D., James J. A., Guthridge J. M., Diamond B., Anolik J. H., Mackey M. F., Alves S. E., Nigrovic P. A., Costenbader K. H., Brenner M. B., Lederer J. A., Rao D. A., Accelerating Medicines Partnership (AMP) RA/SLE Network, PD-1hiCXCR5− T peripheral helper cells promote B cell responses in lupus via MAF and IL-21. JCI Insight 4 (2019).
-
- Makiyama A., Chiba A., Noto D., Murayama G., Yamaji K., Tamura N., Miyake S., Expanded circulating peripheral helper T cells in systemic lupus erythematosus: association with disease activity and B cell differentiation. Rheumatology (Oxford) 58, 1861–1869 (2019). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
