This is a preprint.
Nemo-like kinase disrupts nuclear import and drives TDP43 mislocalization in ALS
- PMID: 39975323
- PMCID: PMC11838369
- DOI: 10.1101/2025.01.27.635090
Nemo-like kinase disrupts nuclear import and drives TDP43 mislocalization in ALS
Update in
-
Nemo-like kinase disrupts nuclear import and drives TDP43 mislocalization in ALS.J Clin Invest. 2025 Jun 24;135(17):e188138. doi: 10.1172/JCI188138. eCollection 2025 Sep 2. J Clin Invest. 2025. PMID: 40553568 Free PMC article.
Abstract
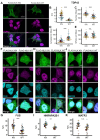
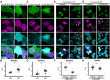
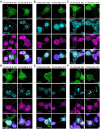
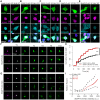
Cytoplasmic TDP43 mislocalization and aggregation are pathological hallmarks of amyotrophic lateral sclerosis (ALS). However, the initial cellular insults that lead to TDP43 mislocalization remain unclear. In this study, we demonstrate that Nemo-like kinase (NLK)-a proline-directed serine/threonine kinase-promotes the mislocalization of TDP43 and other RNA-binding proteins by disrupting nuclear import. NLK levels are selectively elevated in neurons exhibiting TDP43 mislocalization in ALS patient tissues, while genetic reduction of NLK reduces toxicity in human neuron models of ALS. Our findings suggest that NLK is a promising therapeutic target for neurodegenerative diseases.
Conflict of interest statement
S.J.B. serves on the advisory board for Neurocures, Inc., Symbiosis, Eikonizo Therapeutics, 15 Ninesquare Therapeutics, the Live Like Lou Foundation, Synapticure, and the Robert Packard Center for ALS Research. S.J.B. has received research funding from Denali Therapeutics, Biogen, Inc., Lysoway Therapeutics, Amylyx Therapeutics, Acelot Therapeutics, Meira GTX, Inc., Prevail Therapeutics, Eikonizo Therapeutics, and Ninesquare Therapeutics.
Figures


References
-
- Brown RH, and Al-Chalabi A. Amyotrophic Lateral Sclerosis. N Engl J Med. 2017;377(16):1602. - PubMed
-
- Tan RH, Ke YD, Ittner LM, and Halliday GM. ALS/FTLD: experimental models and reality. Acta Neuropathol. 2017;133(2):177–96. - PubMed
-
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–3. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
