This is a preprint.
Identification of actionable targeted protein degradation effector sites through Site-specific Ligand Incorporation-induced Proximity (SLIP)
- PMID: 39975383
- PMCID: PMC11838594
- DOI: 10.1101/2025.02.04.636303
Identification of actionable targeted protein degradation effector sites through Site-specific Ligand Incorporation-induced Proximity (SLIP)
Update in
-
Identification of Actionable Targeted Protein Degradation Effector Sites through Site-Specific Ligand Incorporation-Induced Proximity (SLIP).J Am Chem Soc. 2025 Jun 25;147(25):21549-21559. doi: 10.1021/jacs.5c01420. Epub 2025 Jun 10. J Am Chem Soc. 2025. PMID: 40493711 Free PMC article.
Abstract
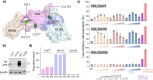
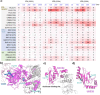
Targeted protein degradation (TPD) is a rapidly emerging and potentially transformative therapeutic modality. However, the large majority of >600 known ubiquitin ligases have yet to be exploited as TPD effectors by proteolysis-targeting chimeras (PROTACs) or molecular glue degraders (MGDs). We report here a chemical-genetic platform, Site-specific Ligand Incorporation-induced Proximity (SLIP), to identify actionable ("PROTACable") sites on any potential effector protein in intact cells. SLIP uses genetic code expansion (GCE) to encode copper-free "click" ligation at a specific effector site in intact cells, enabling in situ formation of a covalent PROTAC-effector conjugate against a target protein of interest (POI). Modification at actionable effector sites drives degradation of the targeted protein, establishing the potential of these sites for TPD. Using SLIP, we systematically screened dozens of sites across E3 ligases and E2 enzymes from diverse classes, identifying multiple novel potentially PROTACable effector sites which are competent for TPD. SLIP adds a powerful approach to the proximity-induced pharmacology (PIP) toolbox, enabling future effector ligand discovery to fully enable TPD, and other emerging PIP modalities.
Keywords: Genetic code expansion; induced proximity; targeted protein degradation.
Figures




References
-
- Popow J.; Farnaby W.; Gollner A.; Kofink C.; Fischer G.; Wurm M.; Zollman D.; Wijaya A.; Mischerikow N.; Hasenoehrl C.; et al. Targeting Cancer with Small-Molecule Pan-KRAS Degraders. Science 2024, 385 (6715), 1338–1347. - PubMed
-
- Tsai J. M.; Nowak R. P.; Ebert B. L.; Fischer E. S. Targeted Protein Degradation: From Mechanisms to Clinic. Nat. Rev. Mol. Cell Biol. 2024, 25 (9), 740–757. - PubMed
-
- Schreiber S. L. Molecular Glues and Bifunctional Compounds: Therapeutic Modalities Based on Induced Proximity. Cell Chem. Biol. 2024, 31 (6), 1050–1063. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
