Application of a new highly multiplexed amplicon sequencing tool to evaluate Plasmodium falciparum antimalarial resistance and relatedness in individual and pooled samples from Dschang, Cameroon
- PMID: 39975479
- PMCID: PMC11835963
- DOI: 10.3389/fpara.2024.1509261
Application of a new highly multiplexed amplicon sequencing tool to evaluate Plasmodium falciparum antimalarial resistance and relatedness in individual and pooled samples from Dschang, Cameroon
Abstract
Background: Resistance to antimalarial drugs remains a major obstacle to malaria elimination. Multiplexed, targeted amplicon sequencing is being adopted for surveilling resistance and dissecting the genetics of complex malaria infections. Moreover, genotyping of parasites and detection of molecular markers drug resistance in resource-limited regions requires open-source protocols for processing samples, using accessible reagents, and rapid methods for processing numerous samples including pooled sequencing.
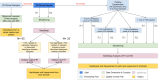
Methods: Plasmodium falciparum Streamlined Multiplex Antimalarial Resistance and Relatedness Testing (Pf-SMARRT) is a PCR-based amplicon panel consisting of 15 amplicons targeting antimalarial resistance mutations and 9 amplicons targeting hypervariable regions. This assay uses oligonucleotide primers in two pools and a non-proprietary library and barcoding approach.
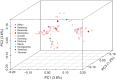
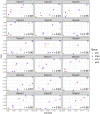
Results: We evaluated Pf-SMARRT using control mocked dried blood spots (DBS) at varying levels of parasitemia and a mixture of 3D7 and Dd2 strains at known frequencies, showing the ability to genotype at low parasite density and recall within-sample allele frequencies. We then piloted Pf-SMARRT to genotype 100 parasite isolates collected from uncomplicated malaria cases at three health facilities in Dschang, Western Cameroon. Antimalarial resistance genotyping showed high levels of sulfadoxine-pyrimethamine resistance mutations, including 31% prevalence of the DHPS A613S mutation. No K13 candidate or validated artemisinin partial resistance mutations were detected, but one low-level non-synonymous change was observed. Pf-SMARRT's hypervariable targets, used to assess complexity of infections and parasite diversity and relatedness, showed similar levels and patterns compared to molecular inversion probe (MIP) sequencing. While there was strong concordance of antimalarial resistance mutations between individual samples and pools, low-frequency variants in the pooled samples were often missed.
Conclusion: Overall, Pf-SMARRT is a robust tool for assessing parasite relatedness and antimalarial drug resistance markers from both individual and pooled samples. Control samples support that accurate genotyping as low as 1 parasite per microliter is routinely possible.
Keywords: Plasmodium falciparum; amplicon sequencing; antimalarial resistance; genetic relatedness; malaria.
Copyright © 2025 Sadler, Simkin, Tchuenkam, Gerdes Gyuricza, Fola, Wamae, Assefa, Niaré, Thwai, White, Moss, Dinglasan, Nsango, Tume, Parr, Ali, Bailey and Juliano.
Conflict of interest statement
JP reports research support from Gilead Sciences, non-financial support from Abbott Laboratories, and consulting for Zymeron Corporation, all outside the scope of the manuscript. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Update of
-
Application of a new highly multiplexed amplicon sequencing tool to evaluate Plasmodium falciparum antimalarial resistance and relatedness in individual and pooled samples from Dschang, Cameroon.medRxiv [Preprint]. 2024 Oct 10:2024.10.03.24314715. doi: 10.1101/2024.10.03.24314715. medRxiv. 2024. Update in: Front Parasitol. 2025 Feb 05;3:1509261. doi: 10.3389/fpara.2024.1509261. PMID: 39417120 Free PMC article. Updated. Preprint.
References
-
- Adegbola A. J., Ijarotimi O. A., Ubom A. E., Adesoji B. A., Babalola O. E., Hocke E. F., et al. . (2023). A snapshot of the prevalence of dihydropteroate synthase-431V mutation and other sulfadoxine-pyrimethamine resistance markers in Plasmodium falciparum isolates in Nigeria. Malar. J. 22, 71. doi: 10.1186/s12936-023-04487-5 - DOI - PMC - PubMed
-
- Ali I. M., Tchuenkam V. P. K., Colton M., Stittleburg V., Mitchell C., Gaither C., et al. . (2022). Arboviruses as an unappreciated cause of non-malarial acute febrile illness in the Dschang Health District of western Cameroon. PloS Negl. Trop. Dis. 16, e0010790. doi: 10.1371/journal.pntd.0010790 - DOI - PMC - PubMed
-
- Aranda-Díaz A., Vickers E. N., Murie K., Palmer B., Hathaway N., Gerlovina I., et al. . (2024). Sensitive and modular amplicon sequencing of Plasmodium falciparum diversity and resistance for research and public health. bioRxiv. 15, 2499.doi: 10.1101/2024.08.22.609145 - DOI
-
- Aydemir O., Janko M., Hathaway N. J., Verity R., Mwandagalirwa M. K., Tshefu A. K., et al. . (2018). Drug-resistance and population structure of plasmodium falciparum across the Democratic Republic of Congo using high-throughput molecular inversion probes. J. Infect. Dis. 218, 946–955. doi: 10.1093/infdis/jiy223 - DOI - PMC - PubMed
-
- Aydemir O., Mensah B., Marsh P. W., Abuaku B., Myers-Hansen J. L., Bailey J. A., et al. . (2021). Immediate pools of malaria infections at diagnosis combined with targeted deep sequencing accurately quantifies frequency of drug resistance mutations. PeerJ 9, e11794. doi: 10.7717/peerj.11794 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

