This is a preprint.
Structural basis of vilazodone dual binding mode to the serotonin transporter
- PMID: 39975905
- PMCID: PMC11838746
- DOI: 10.21203/rs.3.rs-5671197/v1
Structural basis of vilazodone dual binding mode to the serotonin transporter
Abstract
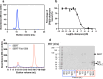
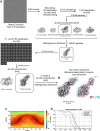
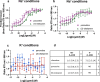
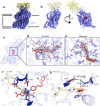
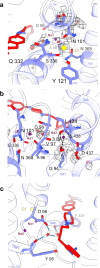
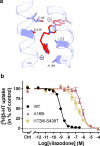
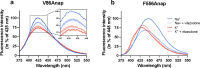
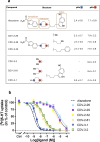
The serotonin transporter (SERT) plays a pivotal role in regulating serotonin (5-HT) signaling and is a key target in treating psychiatric disorders. SERT has a binding site (S1) for 5-HT that also serves as a high-affinity binding site for antidepressants. The antidepressant vilazodone has been shown to inhibit SERT by binding to an allosteric site. Here, we present the cryo-EM structure of SERT with vilazodone bound to the S1 site and extending towards the allosteric site. We systematically dissect the vilazodone molecule into fragments and find that the terminal indole ring is the key determinant for its high affinity to SERT. Further, unlike typical Na+-dependent SERT-selective antidepressants, vilazodone exhibits a dissociation constant (K D) for purified SERT in the nanomolar range both in the presence or absence of Na+. We substantiate this binding mode by exploring the conformational impact of vilazodone binding to SERT using site-specific insertion of the fluorescent non-canonical amino acid Anap. Our results offer novel molecular insight into the distinct pharmacological profile of a clinically used polymodal antidepressant.
Conflict of interest statement
Additional Declarations: There is NO Competing Interest.
Figures








References
-
- World Health Organization. Depression and Other Common Mental Disorders: Global Health Estimates. World Health Organization; (2017).
-
- Garnock-Jones K. P. & McCormack P. L. Escitalopram: A review of its use in the management of major depressive disorder in adults. CNS Drugs 24, 769–796 (2010). - PubMed
-
- Emslie G. J., Mayes T. L. & Ruberu M. Continuation and maintenance therapy of early-onset major depressive disorder. Pediatric Drugs 7, 203–217 (2005). - PubMed
-
- Pundiak T. M., Case B. G., Peselow E. D. & Mulcare L. Discontinuation of maintenance selective serotonin reuptake inhibitor monotherapy after 5 years of stable response: A naturalistic study. J. Clin. Psychiatry 69, 1811–1817 (2008). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

