SIRT3-Mediated Deacetylation of DRP1K711 Prevents Mitochondrial Dysfunction in Parkinson's Disease
- PMID: 39976201
- PMCID: PMC12061286
- DOI: 10.1002/advs.202411235
SIRT3-Mediated Deacetylation of DRP1K711 Prevents Mitochondrial Dysfunction in Parkinson's Disease
Abstract
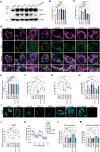
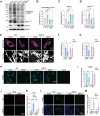
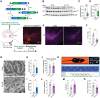
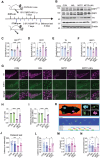
Dysregulation of mitochondrial dynamics is a key contributor to the pathogenesis of Parkinson's disease (PD). Aberrant mitochondrial fission induced by dynamin-related protein 1 (DRP1) causes mitochondrial dysfunction in dopaminergic (DA) neurons. However, the mechanism of DRP1 activation and its role in PD progression remain unclear. In this study, Mass spectrometry analysis is performed and identified a significant increased DRP1 acetylation at lysine residue 711 (K711) in the mitochondria under oxidative stress. Enhanced DRP1K711 acetylation facilitated DRP1 oligomerization, thereby exacerbating mitochondrial fragmentation and compromising the mitochondrial function. DRP1K711 acetylation also affects mitochondrial DRP1 recruitment and fission independent of canonical S616 phosphorylation. Further analysis reveals the critical role of sirtuin (SIRT)-3 in deacetylating DRP1K711, thereby regulating mitochondrial dynamics and function. SIRT3 agonists significantly inhibit DRP1K711 acetylation, rescue DA neuronal loss, and improve motor function in a PD mouse model. Conversely, selective knockout of SIRT3 in DA neurons exacerbates DRP1K711 acetylation, leading to increased DA neuronal damage, neuronal death, and worsened motor dysfunction. Notably, this study identifies a novel mechanism involving aberrant SIRT3-mediated DRP1 acetylation at K711 as a key driver of mitochondrial dysfunction and DA neuronal death in PD, revealing a potential target for PD treatment.
Keywords: DRP1K711; Parkinson's disease; SIRT3; acetylation; mitochondrial dysfunction; oxidative stress.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Ascherio A., Schwarzschild M. A., Lancet Neurol. 2016, 15, 1257. - PubMed
-
- Giannoccaro M. P., La Morgia C., Rizzo G., Carelli V., Mov. Disord. 2017, 32, 346. - PubMed
-
- Poewe W., Seppi K., Tanner C. M., Halliday G. M., Brundin P., Volkmann J., Schrag A. E., Lang A. E., Nat. Rev. Dis. Primers 2017, 3, 17013. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
