Development of ketobenzothiazole-based peptidomimetic TMPRSS13 inhibitors with low nanomolar potency
- PMID: 39976239
- PMCID: PMC11843629
- DOI: 10.1080/14756366.2025.2466841
Development of ketobenzothiazole-based peptidomimetic TMPRSS13 inhibitors with low nanomolar potency
Abstract
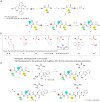
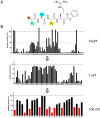
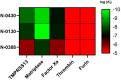
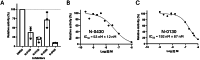
TMPRSS13, a member of the Type II Transmembrane Serine Proteases (TTSP) family, is involved in cancer progression and in respiratory virus cell entry. To date, no inhibitors have been specifically developed for this protease. In this study, a chemical library of 65 ketobenzothiazole-based peptidomimetic molecules was screened against a proteolytically active form of recombinant TMPRSS13 to identify novel inhibitors. Following an initial round of screening, subsequent synthesis of additional derivatives supported by molecular modelling revealed important molecular determinants involved in TMPRSS13 inhibition. One inhibitor, N-0430, achieved low nanomolar affinity towards TMPRSS13 activity in a cellular context. Using a SARS-CoV-2 pseudovirus cell entry model, we further demonstrated the ability of N-0430 to block TMPRSS13-dependent entry of the pseudovirus. The identified peptidomimetic inhibitors and the molecular insights into their potency gained from this study will aid in the development of specific TMPRSS13 inhibitors.
Keywords: Peptidomimetic; SARS-CoV-2; TMPRSS13; compound screening; protease inhibitor.
Figures



Update of
-
Development of ketobenzothiazole-based peptidomimetic TMPRSS13 inhibitors with low nanomolar potency.bioRxiv [Preprint]. 2024 Aug 29:2024.08.28.609965. doi: 10.1101/2024.08.28.609965. bioRxiv. 2024. Update in: J Enzyme Inhib Med Chem. 2025 Dec;40(1):2466841. doi: 10.1080/14756366.2025.2466841. PMID: 39257753 Free PMC article. Updated. Preprint.
Similar articles
-
From N-0385 to N-0920: Unveiling a Host-Directed Protease Inhibitor with Picomolar Antiviral Efficacy against Prevalent SARS-CoV-2 Variants.J Med Chem. 2025 Apr 10;68(7):7119-7136. doi: 10.1021/acs.jmedchem.4c02468. Epub 2025 Mar 31. J Med Chem. 2025. PMID: 40163818
-
Development of ketobenzothiazole-based peptidomimetic TMPRSS13 inhibitors with low nanomolar potency.bioRxiv [Preprint]. 2024 Aug 29:2024.08.28.609965. doi: 10.1101/2024.08.28.609965. bioRxiv. 2024. Update in: J Enzyme Inhib Med Chem. 2025 Dec;40(1):2466841. doi: 10.1080/14756366.2025.2466841. PMID: 39257753 Free PMC article. Updated. Preprint.
-
Optimizing the pharmacokinetics and selectivity of TMPRSS2 inhibitors.Eur J Med Chem. 2025 Sep 15;294:117579. doi: 10.1016/j.ejmech.2025.117579. Epub 2025 Mar 28. Eur J Med Chem. 2025. PMID: 40382841
-
Exploring covalent inhibitors of SARS-CoV-2 main protease: from peptidomimetics to novel scaffolds.J Enzyme Inhib Med Chem. 2025 Dec;40(1):2460045. doi: 10.1080/14756366.2025.2460045. Epub 2025 Feb 6. J Enzyme Inhib Med Chem. 2025. PMID: 39912405 Free PMC article. Review.
-
Recent progress on inhibitors of the type II transmembrane serine proteases, hepsin, matriptase and matriptase-2.Future Med Chem. 2019 Apr;11(7):743-769. doi: 10.4155/fmc-2018-0446. Epub 2019 Apr 4. Future Med Chem. 2019. PMID: 30945556 Review.
Cited by
-
From N-0385 to N-0920: Unveiling a Host-Directed Protease Inhibitor with Picomolar Antiviral Efficacy against Prevalent SARS-CoV-2 Variants.J Med Chem. 2025 Apr 10;68(7):7119-7136. doi: 10.1021/acs.jmedchem.4c02468. Epub 2025 Mar 31. J Med Chem. 2025. PMID: 40163818
References
-
- Webb SL, Sanders AJ, Mason MD, Jiang WG.. Type II transmembrane serine protease (TTSP) deregulation in cancer. Front Biosci. 2011;16(2):539–552. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
