Extracellular Histones as Exosome Membrane Proteins Regulated by Cell Stress
- PMID: 39976275
- PMCID: PMC11840699
- DOI: 10.1002/jev2.70042
Extracellular Histones as Exosome Membrane Proteins Regulated by Cell Stress
Abstract
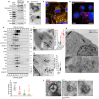
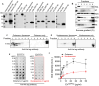
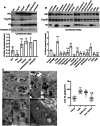
Histones are conserved nuclear proteins that function as part of the nucleosome in the regulation of chromatin structure and gene expression. Interestingly, extracellular histones populate biofluids from healthy individuals, and when elevated, may contribute to various acute and chronic diseases. It is generally assumed that most extracellular histones exist as nucleosomes, as components of extracellular chromatin. We analysed cell culture models under normal and stressed conditions to identify pathways of histone secretion. We report that core and linker histones localize to extracellular vesicles (EVs) and are secreted via the multivesicular body/exosome pathway. Upregulation of EV histone secretion occurs in response to cellular stress, with enhanced vesicle secretion and a shift towards a population of smaller EVs. Most histones were membrane associated with the outer surface of EVs. Degradation of EV-DNA did not impact significantly on EV-histone association. Individual histones and histone octamers bound strongly to liposomes and EVs, but nucleosomes did not, showing histones do not require DNA for EV binding. Histones colocalized to tetraspanin positive EVs but using genetic or pharmacological intervention, we found that all known pathways of exosome biogenesis acted positively on histone secretion. Inhibition of autophagy and lysosomal degradation had a strong positive effect on EV histone release. Unexpectedly, EV-associated histones lacked the extensive post-translational modification of their nuclear counterparts, suggesting loss of PTMs may be involved in their trafficking or secretion. Our data does not support a significant role for EV-histones existing as nucleosomes. We show for the first time that histones are secreted from cells as membrane proteins via EVs/exosomes. This fundamental discovery provides support for further investigation of the biological activity of exosome associated histones and their role in disease.
Keywords: cellular stress; exosome; extracellular vesicles; histone; membrane associated proteins; posttranslational modification.
© 2025 The Author(s). Journal of Extracellular Vesicles published by Wiley Periodicals LLC on behalf of International Society for Extracellular Vesicles.
Conflict of interest statement
Samir El Andaloussi is a founder and consultant for Evox Therapeutics. Samir El Andaloussi, Joel Z. Nordin, and Oscar P. B. Wiklander are shareholders in Evox Therapeutics. The remaining authors declare no competing interests. The other authors declare no conflicts of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

