Trichothiodystrophy due to ERCC2 Variants: Uncommon Contributor to Progressive Hypomyelinating Leukodystrophy
- PMID: 39976384
- PMCID: PMC11840839
- DOI: 10.1002/mgg3.70067
Trichothiodystrophy due to ERCC2 Variants: Uncommon Contributor to Progressive Hypomyelinating Leukodystrophy
Abstract
Background: Trichothiodystrophy (TTD) is caused by homozygous or compound heterozygous variants in genes associated with DNA repair. The ERCC2 gene encoded a protein, XPD, that is a subunit of the general transcription factor TFIIH and important in both DNA repair and transcription. Disease-causing variants in ERCC2 can partially inactivate these activities, giving rise to symptoms seen in TTD, Cockayne syndrome (CS) and xeroderma pigmentosa (XP). Although generalized cerebral white matter abnormalities is reported in TTD, myelination disorders specifically linked to ERCC2 gene variants are exceptionally uncommon. Here, we introduce a thorough investigation of a patient exhibiting classic TTD symptoms alongside progressive cerebral hypomyelination with ERCC2 variants.
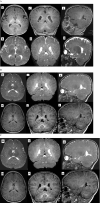
Methods: In a non-consanguineous family, we conducted Autism/ID gene Panel on a 5-year-old affected child who presented with microcephaly, failure to thrive, developmental delay, and progressive hypomyelination on three serial brain imaging over 5-years follow-up. Our investigation aimed to elucidate the genetic underpinnings of the observed phenotype. We also conducted a comprehensive review of the genetic profiles of all documented ERCC2-related patients exhibiting myelination disorders.
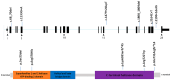
Results: Autism/ID gene Panel identified a compound heterozygous variant in ERCC2 gene causing TTD. Clinical and paraclinical findings enabled differentiation of TTD from Cockayne syndrome and XP. Segregation analysis revealed that, the variation in the paternal allele was a splice junction loss (c.2190 + 1delG), and the other alteration in the maternal allele was a pathogenic variant (c.1479 + 2dupT). It has been noted that these variants were reported in previous studies in homozygous or compound heterozygous form in patients with TTD, but none of them exhibited hypomyelinating leukodystrophy.
Conclusion: The identification of hypomyelination in TTD due to ERCC2 sheds a light on the molecular diagnosis and contributing to the limited literature on ERCC2 variants and associated hypomyelinating leukodystrophy in patients with TTD.
Keywords: ERCC2; hypomyelination; leukodystrophy; trichothiodystrophy.
© 2025 The Author(s). Molecular Genetics & Genomic Medicine published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Persistence of repair proteins at unrepaired DNA damage distinguishes diseases with ERCC2 (XPD) mutations: cancer-prone xeroderma pigmentosum vs. non-cancer-prone trichothiodystrophy.Hum Mutat. 2008 Oct;29(10):1194-208. doi: 10.1002/humu.20768. Hum Mutat. 2008. PMID: 18470933 Free PMC article.
-
Functional and molecular genetic analyses of nine newly identified XPD-deficient patients reveal a novel mutation resulting in TTD as well as in XP/CS complex phenotypes.Exp Dermatol. 2013 Jul;22(7):486-9. doi: 10.1111/exd.12166. Exp Dermatol. 2013. PMID: 23800062
-
Abnormal XPD-induced nuclear receptor transactivation in DNA repair disorders: trichothiodystrophy and xeroderma pigmentosum.Eur J Hum Genet. 2013 Aug;21(8):831-7. doi: 10.1038/ejhg.2012.246. Epub 2012 Dec 12. Eur J Hum Genet. 2013. PMID: 23232694 Free PMC article.
-
Homozygous variants in pyrroline-5-carboxylate reductase 2 (PYCR2) in patients with progressive microcephaly and hypomyelinating leukodystrophy.Am J Med Genet A. 2017 Feb;173(2):460-470. doi: 10.1002/ajmg.a.38049. Epub 2016 Nov 11. Am J Med Genet A. 2017. PMID: 27860360 Review.
-
Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship.Neuroscience. 2007 Apr 14;145(4):1388-96. doi: 10.1016/j.neuroscience.2006.12.020. Epub 2007 Feb 1. Neuroscience. 2007. PMID: 17276014 Free PMC article. Review.
References
-
- Bitoun, E. , Chavanas S., Irvine A. D., et al. 2002. “Netherton Syndrome: Disease Expression and Spectrum of SPINK5 Mutations in 21 Families.” Journal of Investigative Dermatology 118, no. 2: 352–361. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

