Multiple merger coalescent inference of effective population size
- PMID: 39976412
- PMCID: PMC11867189
- DOI: 10.1098/rstb.2023.0306
Multiple merger coalescent inference of effective population size
Abstract
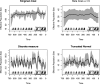
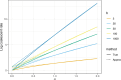
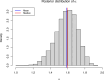
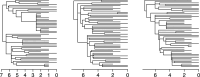
Variation in a sample of molecular sequence data informs about the past evolutionary history of the sample's population. Traditionally, Bayesian modelling coupled with the standard coalescent is used to infer the sample's bifurcating genealogy and demographic and evolutionary parameters such as effective population size and mutation rates. However, there are many situations where binary coalescent models do not accurately reflect the true underlying ancestral processes. Here, we propose a Bayesian non-parametric method for inferring effective population size trajectories from a multifurcating genealogy under the [Formula: see text]-coalescent. In particular, we jointly estimate the effective population size and the model parameter for the Beta-coalescent model, a special type of [Formula: see text]-coalescent. Finally, we test our methods on simulations and apply them to study various viral dynamics as well as Japanese sardine population size changes over time. The code and vignettes can be found in the phylodyn package.This article is part of the theme issue '"A mathematical theory of evolution": phylogenetic models dating back 100 years'.
Keywords: Beta-coalescent; Gaussian processes; Lambda-coalescent; Multiple mergers coalescent.
Conflict of interest statement
We declare we have no competing interests.
Figures


References
-
- Kingman JFC. 1982. The coalescent. Stoch. Process. Their Appl. 13, 235–248. (10.1016/0304-4149(82)90011-4) - DOI
-
- Wakeley J. 2009. Coalescent theory: an introduction. Greenwood Village, CO: Roberts and Co.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

