Transcriptomic landscape of cumulus cells from patients <38 years old with a history of poor ovarian response (POR) treated with platelet-rich plasma (PRP)
- PMID: 39976580
- PMCID: PMC11892918
- DOI: 10.18632/aging.206202
Transcriptomic landscape of cumulus cells from patients <38 years old with a history of poor ovarian response (POR) treated with platelet-rich plasma (PRP)
Abstract
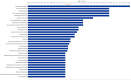
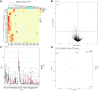
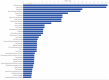
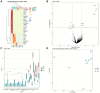
Intraovarian injection of autologous platelet-rich plasma (PRP) has recently been investigated as a potential treatment for patients with diminished ovarian reserve. In the current study, differential gene expression in cumulus cells obtained from patients treated with PRP was compared to controls. RNA sequencing libraries were constructed from the cumulus cells, and differential expression analysis was performed with a false discovery rate threshold of p-value ≤0.05 and Log2 fold change ≥0.584. RNA sequencing of cumulus cells revealed significant differences in gene expression when comparing those treated with PRP and resulted in a live birth (n = 5) to controls with live birth (n = 5), or to controls with failed implantation (n = 5). Similarly, when all samples treated with PRP (those that resulted in live birth or arrested embryos (n = 10)) were compared to all samples from controls (those that resulted in live birth, no pregnancy, or arrested embryos (n = 13)), gene expression was significantly different. Several pathways were consistently affected by PRP treatment through multiple comparisons, including carbohydrate metabolism, cell death and survival, cell growth and proliferation, and cell-to-cell signaling, all of which have been implicated in human causes of infertility.
Keywords: RNA-sequencing; diminished ovarian reserve; plasma rich protein.
Conflict of interest statement
Figures


References
-
- Devine K, Mumford SL, Wu M, DeCherney AH, Hill MJ, Propst A. Diminished ovarian reserve in the United States assisted reproductive technology population: diagnostic trends among 181,536 cycles from the Society for Assisted Reproductive Technology Clinic Outcomes Reporting System. Fertil Steril. 2015; 104:612–9.e3. 10.1016/j.fertnstert.2015.05.017 - DOI - PMC - PubMed
-
- Ferraretti AP, La Marca A, Fauser BC, Tarlatzis B, Nargund G, Gianaroli L, and ESHRE working group on Poor Ovarian Response Definition. ESHRE consensus on the definition of 'poor response' to ovarian stimulation for in vitro fertilization: the Bologna criteria. Hum Reprod. 2011; 26:1616–24. 10.1093/humrep/der092 - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

