miRNome analysis reveals mir-155-5p as a protective factor to dengue infection in a resistant Thai cohort
- PMID: 39976655
- PMCID: PMC11842423
- DOI: 10.1007/s00430-025-00821-7
miRNome analysis reveals mir-155-5p as a protective factor to dengue infection in a resistant Thai cohort
Abstract
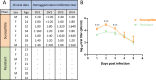
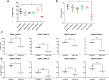
Dengue virus (DENV) is a global health threat, with approximately 390 million infections annually, ranging from mild dengue fever to severe dengue hemorrhagic fever and shock syndrome. MicroRNA (miRNA) are crucial post-transcriptional regulators which may regulate host resistance to DENV infection. This study aimed to identify miRNAs involved in natural resistance to DENV infection. Individuals from a dengue-endemic area were classified as susceptible (SD) or resistant (RD) according to their anti-DENV antibody status. RD individuals were seronegative despite high local DENV infection prevalence. Monocytes susceptibility to DENV infection was assessed in vitro. The miRNome profiles of the monocytes from 7 individuals per group were assessed upon mock or DENV-2 infection. The antiviral effect of differentially expressed miRNAs was analyzed using miRNA mimics in HeLa cells followed by infection with DENV-1, DENV-2, DENV-3, and DENV-4 serotypes. We performed RNA-seq on miRNA mimic-transfected cells to identify miRNA-targeted genes interacting with DENV proteins. Monocytes from RD individuals exhibit lower DENV-2 production in vitro. The miRNAs miR-155, miR-132-3p, miR-576-5p were overexpressed in monocytes from RD group upon DENV-2 infection. The transfection of miR-155-5p mimic reduced DENV infection and viral production in HeLa cells, regulating 18 genes interacting with DENV proteins and downregulating target genes involved in interferon response, TP53 regulation, apoptosis, and vesicle trafficking (e.g. HSD17B12, ANXA2). Therefore, we show that monocytes from RD individuals show a distinct miRNA expression profile and reduced viral production. In vitro miR-155-5p upregulation induces an antiviral state, revealing potential therapeutic targets to treat dengue.
Keywords: Dengue; RNA-Seq; Resistance; miR-155-5p; miRNome; microRNA.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: Ethical approval was received by the Medical Ethics Committee of the Mahidol University, Thailand under registration number COA N° MU-IRB 2012/180.1311. A written informed consent was obtained from the volunteers before the blood samples collection. Competing interests: The authors declare no competing interests.
Figures


References
-
- Khan MB, Yang Z-S, Lin C-Y et al (2023) Dengue overview: an updated systemic review. J Infect Public Health 16:1625–1642. 10.1016/j.jiph.2023.08.001 - PubMed
-
- van der Ree MH, van der Meer AJ, de Bruijne J et al (2014) Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res 111:53–59. 10.1016/j.antiviral.2014.08.015 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

