Enterobacter hormaechei replaces virulence with carbapenem resistance via porin loss
- PMID: 39977318
- PMCID: PMC11874173
- DOI: 10.1073/pnas.2414315122
Enterobacter hormaechei replaces virulence with carbapenem resistance via porin loss
Abstract
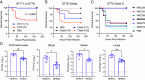
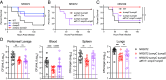
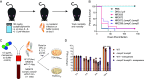
Pathogenic Enterobacter species are of increasing clinical concern due to the multidrug-resistant nature of these bacteria, including resistance to carbapenem antibiotics. Our understanding of Enterobacter virulence is limited, hindering the development of new prophylactics and therapeutics targeting infections caused by Enterobacter species. In this study, we assessed the virulence of contemporary clinical Enterobacter hormaechei isolates in a mouse model of intraperitoneal infection and used comparative genomics to identify genes promoting virulence. Through mutagenesis and complementation studies, we found two porin-encoding genes, ompC and ompD, to be required for E. hormaechei virulence. These porins imported clinically relevant carbapenems into the bacteria, and thus loss of OmpC and OmpD desensitized E. hormaechei to the antibiotics. Our genomic analyses suggest porin-related genes are frequently mutated in E. hormaechei, perhaps due to the selective pressure of antibiotic therapy during infection. Despite the importance of OmpC and OmpD during infection of immunocompetent hosts, we found the two porins to be dispensable for virulence in a neutropenic mouse model. Moreover, porin loss provided a fitness advantage during carbapenem treatment in an ex vivo human whole blood model of bacteremia. Our data provide experimental evidence of pathogenic Enterobacter species gaining antibiotic resistance via loss of porins and argue antibiotic therapy during infection of immunocompromised patients is a conducive environment for the selection of porin mutations enhancing the multidrug-resistant profile of these pathogens.
Keywords: Enterobacter cloacae complex; antimicrobial resistance; infection and pathogenesis; porins; virulence.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


References
-
- Souza Lopes A. C., et al. , Occurrence and analysis of irp2 virulence gene in isolates of Klebsiella pneumoniae and Enterobacter spp. from microbiota and hospital and community-acquired infections. Microb. Pathog. 96, 15–19 (2016). - PubMed
-
- Bousquet A., et al. , Outbreak of CTX-M-15-producing Enterobacter cloacae associated with therapeutic beds and syphons in an intensive care unit. Am. J. Infect. Control 45, 1160–1164 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

