Molecular Analysis of Human Respiratory Syncytial Virus Group B Strains Isolated in Kenya Before and During the Emergence of Pandemic Influenza A/H1N1
- PMID: 39978327
- PMCID: PMC11842092
- DOI: 10.1111/irv.70082
Molecular Analysis of Human Respiratory Syncytial Virus Group B Strains Isolated in Kenya Before and During the Emergence of Pandemic Influenza A/H1N1
Abstract
Background: We conducted a retrospective study to explore molecular insights into human respiratory syncytial virus (HRSV) group B strains among patients attending outpatient clinics at government medical facilities both prior and during the onset of Influenza A/H1N1/2009 pandemic outbreak.
Methods: We screened 2300 nasopharyngeal swabs using multiplex real time reverse transcriptase polymerase chain reaction. We amplified a segment of the first and second hypervariable regions, as well as the conserved portion of the third domain of the G-gene using HRSV-B specific primers, sequenced by Sanger di-deoxy chain termination method and thereafter analyzed the sequences.
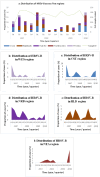
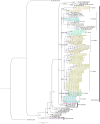
Results: We characterized the circulating strains into three known genotypes: SAB4 (1.4%), BA7 (1.4%), and multiple variants of BA9 (97.2%). The majority of BA9 viruses were uniquely Kenyan with only 4% aligning with BA9 lineages found elsewhere. The mean evolutionary rate of the HRSV-B was estimated to be 3.08 × 10-3 substitutions per site per year.
Conclusion: Our findings indicate that the circulating HRSV-B viruses in Kenya underwent a slower evolution during the period of 2007-2010. Additionally, our findings reveal the existence of a unique lineage as well as new variants that have not been reported elsewhere to date.
Keywords: COVID‐19 pandemic; HRSV‐B genotypes; SARS‐CoV‐2; evolution; genetic variability; pandemic influenza; recombination; spatial–temporal trends.
© 2025 The Author(s). Influenza and Other Respiratory Viruses published by John Wiley & Sons Ltd.
Conflict of interest statement
The views or insertions expressed herein are private views of the authors and are not to be construed to represent those of the US Department of Defense or Army.
The authors declare no conflicts of interest.
Figures


Similar articles
-
Genetic variation of human respiratory syncytial virus among children with fever and respiratory symptoms in Shanghai, China, from 2009 to 2012.Infect Genet Evol. 2014 Oct;27:131-6. doi: 10.1016/j.meegid.2014.07.011. Epub 2014 Jul 19. Infect Genet Evol. 2014. PMID: 25046173
-
Molecular characterization and phylogenetic analysis of the hemagglutinin 1 protein of human influenza A virus subtype H1N1 circulating in Kenya during 2007-2008.J Infect Dis. 2012 Dec 15;206 Suppl 1:S46-52. doi: 10.1093/infdis/jis586. J Infect Dis. 2012. PMID: 23169971
-
Molecular characterization of human respiratory syncytial virus subtype B: a novel genotype of subtype B circulating in China.J Med Virol. 2015 Jan;87(1):1-9. doi: 10.1002/jmv.23960. Epub 2014 Jun 9. J Med Virol. 2015. PMID: 24910250
-
Prevalence and genetic characterisation of respiratory syncytial viruses circulating in Bulgaria during the 2014/15 and 2015/16 winter seasons.Pathog Glob Health. 2017 Oct;111(7):351-361. doi: 10.1080/20477724.2017.1375708. Epub 2017 Sep 26. Pathog Glob Health. 2017. PMID: 28948867 Free PMC article.
-
Whole-Genome Sequencing and Genetic Diversity of Human Respiratory Syncytial Virus in Patients with Influenza-like Illness in Sicily (Italy) from 2017 to 2023.Viruses. 2024 May 26;16(6):851. doi: 10.3390/v16060851. Viruses. 2024. PMID: 38932144 Free PMC article.
Cited by
-
Development and Validation of a New Set of Primers for Identification of Circulating Lineages and Palivizumab/Nirsevimab Resistance in HRSV Isolates from Cabo Verde.Trop Med Infect Dis. 2025 Jun 10;10(6):160. doi: 10.3390/tropicalmed10060160. Trop Med Infect Dis. 2025. PMID: 40559727 Free PMC article.
References
-
- Peret T. C., Hall C. B., Hammond G. W., et al., “Circulation Patterns of Group A and B Human Respiratory Syncytial Virus Genotypes in 5 Communities in North America,” Journal of Infectious Diseases 181 (2000): 1891–1896. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

