Deciphering response dynamics and treatment resistance from circulating tumor DNA after CAR T-cells in multiple myeloma
- PMID: 39979252
- PMCID: PMC11842827
- DOI: 10.1038/s41467-025-56486-6
Deciphering response dynamics and treatment resistance from circulating tumor DNA after CAR T-cells in multiple myeloma
Abstract
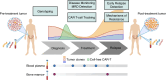
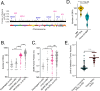
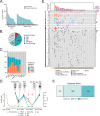
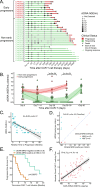
Despite advances in treatments, multiple myeloma (MM) remains an incurable cancer where relapse is common. We developed a circulating tumor DNA (ctDNA) approach in order to characterize tumor genomics, monitor treatment response, and detect early relapse in MM. By sequencing 412 specimens from 64 patients with newly diagnosed or relapsed/refractory disease, we demonstrate the correlation between ctDNA and key clinical biomarkers, as well as patient outcomes. We further extend our approach to simultaneously track CAR-specific cell-free DNA (CAR-cfDNA) in patients undergoing anti-BCMA CAR T-cell (BCMA-CAR) therapy. We demonstrate that ctDNA levels following BCMA-CAR inversely correlate with relative time to progression (TTP), and that measurable residual disease (MRD) quantified by peripheral blood ctDNA (ctDNA-MRD) was concordant with clinical bone marrow MRD. Finally, we show that ctDNA-MRD can anticipate clinical relapse and identify the emergence of genomically-defined therapy-resistant clones. These findings suggest multiple clinical uses of ctDNA for MM in molecular characterization and disease surveillance.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: B.J.S.: consultancy: Foresight Diagnostics. M.L.: advisory board/consulting for Janssen, BMS, and Kite. D.M.: patent: Pharmacyclics supporting ibrutinib for chronic graft-vs-host disease. Research funding and consultancy: Pharmacyclics, Kite Pharma, Adaptive Biotechnologies, Novartis, Juno Therapeutics, Celgene, Janssen Pharmaceuticals, Roche, Genentech, Precision Bioscience, Allogene and Miltenyi Biotec. M.S.K: research funding: Nutcracker Therapeutics, CRISPR Therapeutics; Consultancy: Daiichi Sankyo, Myeloid Therapeutics. S.S.: research funding: Magenta Therapeutics, BMS, Allogene, Janssen; Consultancy: Magenta Therapeutics, BMS, Janssen, Sanofi, Oncopeptides, Takeda. D.M.K.: patents: D.M.K. reports issued patents pertaining to ctDNA-MRD detection assigned to Stanford University and licensed to Foresight Diagnostics. Consultancy: Foresight Diagnostics, Roche, Genentech, and Adaptive Biotechnologies. Stock and Equity Ownership: D.M.K. reports equity ownership in Foresight Diagnostics. The remaining authors declare no competing interests.
Figures

References
-
- Braunlin, M., Belani, R., Buchanan, J., Wheeling, T. & Kim, C. Trends in the multiple myeloma treatment landscape and survival: a U.S. analysis using 2011–2019 oncology clinic electronic health record data. Leuk. Lymphoma62, 377–386 (2021). - PubMed
-
- Binder, M. et al. Mortality trends in multiple myeloma after the introduction of novel therapies in the United States. Leukemia36, 801–808 (2022). - PubMed
-
- Corre, J. et al. Improved survival in multiple myeloma during the 2005–2009 and 2010–2014 periods. Leukemia35, 3600–3603 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

