Tubular CD44 plays a key role in aggravating AKI through NF-κB p65-mediated mitochondrial dysfunction
- PMID: 39979265
- PMCID: PMC11842857
- DOI: 10.1038/s41419-025-07438-x
Tubular CD44 plays a key role in aggravating AKI through NF-κB p65-mediated mitochondrial dysfunction
Abstract
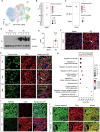
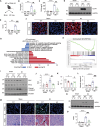
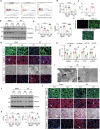
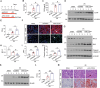
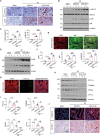
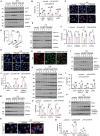
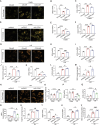
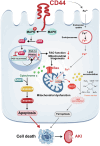
Acute kidney injury (AKI) is in rapid prevalence nowadays. Of note, the underlying mechanisms have not been clarified. Several reports showed a cluster of differentiation-44 (CD44), a cell-surface glycoprotein, might be involved in AKI. However, its role in AKI has not been clearly clarified. Herein, we found CD44 increased in renal tubules in AKI mice. Gene ablation of CD44 improved mitochondrial biogenesis and fatty acid oxidation (FAO) function, further protecting against tubular cell death and kidney injury. Conversely, ectopic CD44 impaired mitochondrial homeostasis and exacerbated tubular cell apoptosis to aggravate AKI progression. From transcriptome sequencing, we found that CD44 induces mitogen-activated protein kinase (MAPK) and NF-κB p65 signaling. Lipidomics also showed that CD44 interfered with multiple aspects of lipid metabolism. We deeply investigated NF-κB p65 inhibited the transcription of peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), resulting in mitochondrial dysfunction and cell apoptosis. CD44 also facilitated iron intake to assist cell ferroptosis. Hence, our study provided a new mechanism for AKI, and demonstrated that targeted inhibition on CD44 could be a promising therapeutic strategy to resist AKI.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics statement: The animal experiments were approved by the Ethics Committee on Use and Care of Animals of Southern Medical University, Guangzhou, China.
Figures


Similar articles
-
DNAJC6 in acute kidney injury: A novel target for protecting renal tubular epithelial cells through PGC-1α-mediated mitochondrial homeostasis.Exp Cell Res. 2025 Jul 15;450(2):114682. doi: 10.1016/j.yexcr.2025.114682. Epub 2025 Jul 21. Exp Cell Res. 2025. PMID: 40701388
-
PGC-1α deficiency causes spontaneous kidney inflammation and increases the severity of nephrotoxic AKI.J Pathol. 2019 Sep;249(1):65-78. doi: 10.1002/path.5282. Epub 2019 Jun 12. J Pathol. 2019. PMID: 30982966
-
Effect of forkhead box O1 in renal tubular epithelial cells on endotoxin-induced acute kidney injury.Am J Physiol Renal Physiol. 2021 Mar 1;320(3):F262-F272. doi: 10.1152/ajprenal.00289.2020. Epub 2020 Dec 28. Am J Physiol Renal Physiol. 2021. PMID: 33356954
-
The Role of PGC-1α and Mitochondrial Biogenesis in Kidney Diseases.Biomolecules. 2020 Feb 24;10(2):347. doi: 10.3390/biom10020347. Biomolecules. 2020. PMID: 32102312 Free PMC article. Review.
-
The Role of Peroxisome Proliferator-Activated Receptor γ Coactivator 1α (PGC-1α) in Kidney Disease.Semin Nephrol. 2018 Mar;38(2):121-126. doi: 10.1016/j.semnephrol.2018.01.003. Semin Nephrol. 2018. PMID: 29602395 Free PMC article. Review.
References
-
- Singbartl K, Kellum JA. AKI in the ICU: definition, epidemiology, risk stratification, and outcomes. Kidney Int. 2012;81:819–25. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

