Peroxisomal core structures segregate diverse metabolic pathways
- PMID: 39979331
- PMCID: PMC11842775
- DOI: 10.1038/s41467-025-57053-9
Peroxisomal core structures segregate diverse metabolic pathways
Abstract
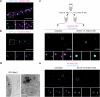
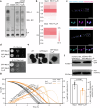
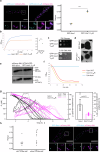
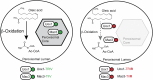
Peroxisomes are single membrane-bounded oxidative organelles with various metabolic functions including β-oxidation of fatty acids. Peroxisomes of many species confine certain metabolic enzymes into sub-compartments sometimes visible as electron dense cores. Why these structures form is largely unknown. Here, we report that in the smut fungus Ustilago maydis detergent resistant core structures are enriched for different enzymes excluding several key enzymes of the β-oxidation pathway. This confinement contributes to generation of peroxisome subpopulations that differ in their enzyme content. We identify short amino acid motifs necessary and sufficient for protein self-assembly into aggregates in vitro. The motifs trigger enrichment in cores in vivo and are active in mammalian cells. Perturbation of core assembly via variation of such motifs affects peroxisome function in U. maydis strains challenged with fatty acids. Thus, protein core structures serve to compartmentalize the lumen of peroxisomes thereby preventing interference of biochemical reactions. Metabolic compartmentalization of peroxisomes via assembly of specific proteins may occur in other organisms as well.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Poirier, Y., Antonenkov, V. D., Glumoff, T. & Hiltunen, J. K. Peroxisomal β-oxidation—a metabolic pathway with multiple functions. Biochim. Biophys. Acta1763, 1413–1426 (2006). - PubMed
-
- Stehlik, T., Sandrock, B., Ast, J. & Freitag, J. Fungal peroxisomes as biosynthetic organelles. Curr. Opin. Microbiol.22, 8–14 (2014). - PubMed
-
- Fujiki, Y., Okumoto, K., Honsho, M. & Abe, Y. Molecular insights into peroxisome homeostasis and peroxisome biogenesis disorders. Biochim. Biophys. Acta Mol. Cell Res.1869, 119330 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

