Genetically encoded biosensor for fluorescence lifetime imaging of PTEN dynamics in the intact brain
- PMID: 39979596
- PMCID: PMC11978514
- DOI: 10.1038/s41592-025-02610-9
Genetically encoded biosensor for fluorescence lifetime imaging of PTEN dynamics in the intact brain
Abstract
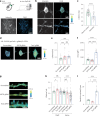
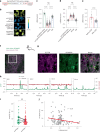
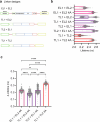
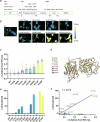
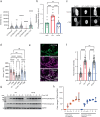
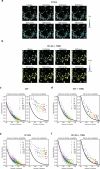
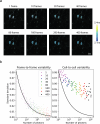
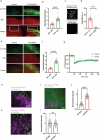
The phosphatase and tensin homolog (PTEN) is a vital protein that maintains an inhibitory brake for cellular proliferation and growth. Accordingly, PTEN loss-of-function mutations are associated with a broad spectrum of human pathologies. Despite its importance, there is currently no method to directly monitor PTEN activity with cellular specificity within intact biological systems. Here we describe the development of a FRET-based biosensor using PTEN conformation as a proxy for the PTEN activity state, for two-photon fluorescence lifetime imaging microscopy. We identify a point mutation that allows the monitoring of PTEN activity with minimal interference to endogenous PTEN signaling. We demonstrate imaging of PTEN activity in cell lines, intact Caenorhabditis elegans and in the mouse brain. Finally, we develop a red-shifted sensor variant that allows us to identify cell-type-specific PTEN activity in excitatory and inhibitory cortical cells. In summary, our approach enables dynamic imaging of PTEN activity in vivo with unprecedented spatial and temporal resolution.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







References
-
- Worby, C. A. & Dixon, J. E. PTEN. Annu. Rev. Biochem.83, 641–669 (2014). - PubMed
-
- Stambolic, V. et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell95, 29–39 (1998). - PubMed
-
- Funamoto, S., Meili, R., Lee, S., Parry, L. & Firtel, R. A. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell109, 611–623 (2002). - PubMed
-
- Li, Z. et al. Regulation of PTEN by Rho small GTPases. Nat. Cell Biol.7, 399–404 (2005). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

