Metabolic engineering of Komagataella phaffii for enhanced 3-hydroxypropionic acid (3-HP) production from methanol
- PMID: 39979934
- PMCID: PMC11844118
- DOI: 10.1186/s13036-025-00488-x
Metabolic engineering of Komagataella phaffii for enhanced 3-hydroxypropionic acid (3-HP) production from methanol
Abstract
Background: Bioconversion of methanol derived from CO2 reduction into value-added chemicals provides a unique approach for mitigating global warming and reducing fossil fuels dependence. Production of 3-hydroxypropionic acid (3-HP), a key building block for the development of biobased products such as acrylates and 1,3-propanediol, has been successfully achieved using methanol as the sole carbon and energy source in the methylotrophic yeast Komagataella phaffii (syn. Pichia pastoris). However, challenges remain in meeting commercially relevant concentrations, yields and productivities of 3-HP, prompting further strain optimization. In the present study, we have combined metabolic engineering strategies aiming at increasing metabolic precursors supply and redirecting carbon flux towards 3-HP production.
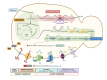
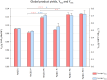
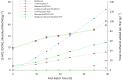
Results: A combinatorial metabolic engineering strategy targeting precursors supply and 3-HP export was applied to the original 3-HP producing K. phaffii strain harboring the synthetic β-alanine pathway and a mutated NADP-dependent formate dehydrogenase from Pseudomonas sp. 101 (PseFDH(V9)). To do so, several genes encoding enzymes catalyzing reactions immediately upstream of the β-alanine pathway were overexpressed to enhance precursors availability. However, only the overexpression of the pyruvate carboxylase PYC2 gene significantly increased the 3-HP yield on biomass (YP/X) in small-scale cultivations. Co-overexpression of PYC2 and the lactate permeases ESBP6 and JEN1 genes led to a 55% improvement in 3-HP titer and product yield in methanol deep-well plate cultures compared to the reference strain, mostly due to Esbp6 activity, proving its effectiveness as a 3-HP transporter. Deletion of the native formate dehydrogenase gene FDH1 did not increase methanol flux entering the assimilatory pathway. Instead, knockout strains showed severe growth defects due to toxic intermediates accumulation. Co-expression of the PseFDH(V9) encoding gene in these strains failed to compensate for the loss of the native FDH. The strain combining PYC2, ESBP6, and JEN1 overexpression was further tested in fed-batch cultures at pH 5, achieving a 3-HP concentration of 27.0 g l- 1, with a product yield of 0.19 g g- 1, and a volumetric productivity of 0.56 g l- 1 h- 1 for the methanol feeding phase of the cultivations. These results represent a 42% increase in final concentration and over 20% improvement in volumetric productivity compared to the original 3-HP-producing strain. Furthermore, bioreactor-scale cultivations at pH 3.5 revealed increased robustness of the strains overexpressing monocarboxylate transporters.
Conclusions: Our results point out the potential of lactate transporters to efficiently drive 3-HP export in K. phaffii, leading to higher titers, yields, and productivities, even at lower pH conditions.
Keywords: Komagataella phaffii; Pichia pastoris; 3-hydroxypropionic acid; Lactate transporters; Metabolic engineering; Methanol; ß-alanine pathway.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

