Genotyping-by-sequencing derived SNP markers reveal genetic diversity and population structure of Dactylis glomerata germplasm
- PMID: 39980483
- PMCID: PMC11840758
- DOI: 10.3389/fpls.2025.1530585
Genotyping-by-sequencing derived SNP markers reveal genetic diversity and population structure of Dactylis glomerata germplasm
Abstract
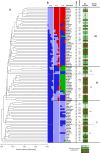
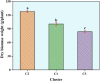
Orchardgrass (Dactylis glomerata L.), a widely cultivated cool-season perennial, is an important forage crop due to its adaptability, high nutritional value, and substantial biomass. Understanding its genetic diversity and population structure is crucial for developing resilient cultivars that can withstand climate change, diseases, and resource limitations. Despite its global significance in fodder production, the genetic potential of many regional accessions remains unexplored, limiting breeding efforts. This study investigates the genetic diversity (GD) and population structure of 91 accessions of D. glomerata from Turkey and Iran using genotyping-by-sequencing based single nucleotide polymorphism (SNP) markers. A total of 2913 high-quality SNP markers revealed substantial genetic variability across provinces. Notably, accessions from Erzurum exhibited the highest GD (mean GD: 0.26; He: 0.5328), while provinces such as Bursa and Muğla demonstrated lower GD (mean GD: 0.15; He < 0.22), suggesting potential genetic bottlenecks. Population structure analysis using Bayesian clustering, PCoA and UPGMA dendrograms divided the accessions into three distinct clusters, with cluster membership largely reflecting geographical origins, and dry biomass content. Cluster II revealed higher GD, associated with enhanced biomass production (128 g/plant), the most important agronomic trait in forage species, supporting the notion of heterosis in breeding programs. The majority of the genetic variation (85.8%) was observed within clusters, with minimal differentiation among clusters (FST = 0.007). Genome-wide association studies (GWAS) identified significant marker-trait associations for dry biomass weight, a critical agronomic trait, with markers DArT-100715788, DArT-101043591, and DArT-101171265 and DArT-101090822 located on Chromosomes 1, 6, and 7 respectively. These findings highlight the importance of regional diversity for maintaining adaptive potential in future breeding programs.
Keywords: Dactylis glomerata; GWAS; SNP markers; forage breeding; genetic diversity; population structure.
Copyright © 2025 Altaf, Cavagnaro, Kökten, Ali, Morales, Tatar, Bedir, Nadeem, Aasim, Çeliktaş, Mansoor and Baloch.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Altaf M. T., Liaqat W., Ali A., Jamil A., Bedir M., Nadeem M. A., et al. . (2024. b). “Conventional and Biotechnological Approaches for the Improvement of Industrial Crops,” in Industrial Crop Plants (Springer Nature Singapore, Singapore: ), 1–48. doi: 10.1007/978-981-97-1003-4_1 - DOI
-
- Altaf M. T., Nadeem M. A., Ali A., Liaqat W., Bedir M., Baran N., et al. . (2024. a). Applicability of Start Codon Targeted (SCoT) markers for the assessment of genetic diversity in bread wheat germplasm. Genet. Resour. Crop Evol., 1–14. doi: 10.1007/s10722-024-02016-0 - DOI
-
- Altaf M. T., Nadeem M. A., Ayten S., Aksoy E., Sönmez F., Çiftçi V., et al. . (2024. d). Unveiling genetic basis associated with manganese content in Turkish common bean (Phaseolus vulgaris L.) germplasm through a genome-wide association study. Plant Breed., 1–25. doi: 10.1111/pbr.13251 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

