Impaired inflammatory resolution with severe SARS-CoV-2 infection in leptin knock out obese hamster
- PMID: 39981511
- PMCID: PMC11841202
- DOI: 10.1016/j.isci.2025.111837
Impaired inflammatory resolution with severe SARS-CoV-2 infection in leptin knock out obese hamster
Abstract
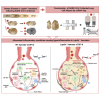
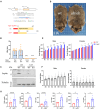
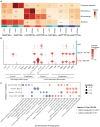
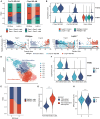
Comorbidities, such as obesity, increase the risk of severe COVID-19. However, the mechanisms underlying severe illnesses in individuals with obesity are poorly understood. Here, we used gene-edited leptin knock out (Leptin -/-) obese hamsters to establish a severe infection model. This model exhibits robust viral replication, severe lung lesions, pronounced clinical symptoms, and fatal infection, mirroring severe COVID-19 in patients with obesity. Using single-cell transcriptomics on lung tissues pre- and post-infection, we found that monocyte-derived alveolar macrophages (MD-AM) play a key role in lung hyper-inflammation, including two unique MD-AM cell fate branches specific to Leptin -/- hamsters. Notably, reduced Trem2-dependent efferocytosis pathways in Leptin -/- hamsters indicated weakened inflammation resolution, consistent with the scRNA-seq data from patients with obesity. In summary, our study highlights the obesity-associated mechanisms underlying severe SARS-CoV-2 infections and establishes a reliable preclinical animal model for developing obesity-specific therapeutics for critical COVID-19.
Keywords: Immune response; Omics; Virology.
© 2025 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Banerjee A., Pasea L., Harris S., Gonzalez-Izquierdo A., Torralbo A., Shallcross L., Noursadeghi M., Pillay D., Sebire N., Holmes C., et al. Estimating excess 1-year mortality associated with the COVID-19 pandemic according to underlying conditions and age: a population-based cohort study. Lancet. 2020;395:1715–1725. doi: 10.1016/S0140-6736(20)30854-0. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

