Spatial Radiomic Graphs for Outcome Prediction in Radiation Therapy-treated Head and Neck Squamous Cell Carcinoma Using Pretreatment CT
- PMID: 39982207
- PMCID: PMC11966549
- DOI: 10.1148/rycan.240161
Spatial Radiomic Graphs for Outcome Prediction in Radiation Therapy-treated Head and Neck Squamous Cell Carcinoma Using Pretreatment CT
Abstract
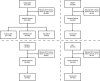
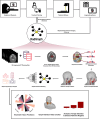
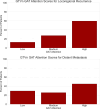
Purpose To develop a radiomic graph framework, RadGraph, for spatial analysis of pretreatment CT images to improve prediction of local-regional recurrence (LR) and distant metastasis (DM) in head and neck squamous cell carcinoma (HNSCC). Materials and Methods This retrospective study included four public pre-radiotherapy treatment CT datasets of patients with HNSCC obtained from The Cancer Imaging Archive (images collected between 2003 and 2018). Computational graphs and graph attention deep learning methods were leveraged to holistically model multiple regions in the head and neck anatomy. Clinical features, including age, sex, and human papillomavirus infection status, were collected for a baseline model. Model performance in predicting LR and DM was evaluated via area under the receiver operating characteristic curve (AUC) and qualitative interpretation of model attention. Results A total of 3434 patients (61 years ± 11 [SD], 2774 male) were divided into training (n = 1576), validation (n = 379), and testing (n = 1479) datasets. RadGraph achieved AUCs of up to 0.83 and 0.90 for LR and DM prediction, respectively. RadGraph showed higher performance compared with the clinical baseline (AUCs up to 0.73 for LR prediction and 0.83 for DM prediction) and previously published approaches (AUCs up to 0.81 for LR prediction and 0.87 for DM prediction). Graph attention atlases enabled visualization of regions coinciding with cervical lymph node chains as important for outcome prediction. Conclusion RadGraph leveraged information from tumor and nontumor regions to effectively predict LR and DM in a large multi-institutional dataset of patients with radiation therapy-treated HNSCC. Graph attention atlases enabled interpretation of model predictions. Keywords: CT, Informatics, Neural Networks, Radiation Therapy, Head/Neck, Computer Applications-General (Informatics), Tumor Response, Head and Neck Squamous Cell Carcinoma, Locoregional Recurrence, Radiotherapy, Deep Learning, Radiomics Supplemental material is available for this article. © RSNA, 2025.
Keywords: CT; Computer Applications–General (Informatics); Deep Learning; Head and Neck Squamous Cell Carcinoma; Head/Neck; Informatics; Locoregional Recurrence; Neural Networks; Radiation Therapy; Radiomics; Radiotherapy; Tumor Response.
Conflict of interest statement
Figures



References
-
- Taneja C , Allen H , Koness RJ , Radie-Keane K , Wanebo HJ . Changing patterns of failure of head and neck cancer . Arch Otolaryngol Head Neck Surg 2002. ; 128 ( 3 ): 324 – 327 . - PubMed
-
- Fischer CA , Kampmann M , Zlobec I , et al. . p16 expression in oropharyngeal cancer: its impact on staging and prognosis compared with the conventional clinical staging parameters . Ann Oncol 2010. ; 21 ( 10 ): 1961 – 1966 . - PubMed
-
- McDuffie CM , Amirghahari N , Caldito G , Lian TS , Thompson L , Nathan CO . Predictive factors for posterior triangle metastasis in HNSCC . Laryngoscope 2005. ; 115 ( 12 ): 2114 – 2117 . - PubMed
-
- Ferlito A , Rinaldo A , Devaney KO , et al. . Prognostic significance of microscopic and macroscopic extracapsular spread from metastatic tumor in the cervical lymph nodes . Oral Oncol 2002. ; 38 ( 8 ): 747 – 751 . - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

