Spatio-temporal transcriptomic analysis reveals distinct nephrotoxicity, DNA damage, and regeneration response after cisplatin
- PMID: 39982567
- PMCID: PMC11845422
- DOI: 10.1007/s10565-025-10003-z
Spatio-temporal transcriptomic analysis reveals distinct nephrotoxicity, DNA damage, and regeneration response after cisplatin
Abstract
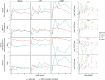
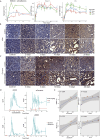
Nephrotoxicity caused by drug or chemical exposure involves complex mechanisms as well as a temporal integration of injury and repair responses in different nephron segments. Distinct cellular transcriptional programs regulate the time-dependent tissue injury and regeneration responses. Whole kidney transcriptome analysis cannot dissect the complex spatio-temporal injury and regeneration responses in the different nephron segments. Here, we used laser capture microdissection of formalin-fixed paraffin embedded sections followed by whole genome targeted RNA-sequencing-TempO-Seq and co-expression gene-network (module) analysis to determine the spatial-temporal responses in rat kidney glomeruli (GM), cortical proximal tubules (CPT) and outer-medulla proximal tubules (OMPT) comparison with whole kidney, after a single dose of the nephrotoxicant cisplatin. We demonstrate that cisplatin induced early onset of DNA damage in both CPT and OMPT, but not GM. Sustained DNA damage response was strongest in OMPT coinciding with OMPT specific inflammatory signaling, actin cytoskeletal remodeling and increased glycolytic metabolism with suppression of mitochondrial activity. Later responses reflected regeneration-related cell cycle pathway activation and ribosomal biogenesis in the injured OMPT regions. Activation of modules containing kidney injury biomarkers was strongest in OMPT, with OMPT Clu expression highly correlating with urinary clusterin biomarker measurements compared the correlation of Kim1. Our findings also showed that whole kidney responses were less sensitive than OMPT. In conclusion, our LCM-TempO-Seq method reveals a detailed spatial mechanistic understanding of renal injury/regeneration after nephrotoxicant exposure and identifies the most representative mechanism-based nephron segment specific renal injury biomarkers.
Keywords: Cisplatin; Nephrons; Nephrotoxicity; Spatial; Temporal; Transcriptomics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: Both animal dosing studies were performed at AbbVie Deutschland GmbH & Co. KG (Knollstrasse, 67061 Ludwigshafen, Germany) in accordance with the German Federal Animal Welfare Act. Conflict of interest: M.E.C, currently employed at Regeneron Pharmaceuticals Inc, contributed to this article as an employee of Abbvie and the views expressed do not necessarily represent the views of Regeneron Pharmaceuticals Inc. P.T, currently employed at Sanofi, contributed to this article as an employee of Abbvie and the views expressed do not necessarily represent the views of Sanofi.
Figures







References
-
- Benedetti G, Fredriksson L, Herpers B, Meerman J, van de Water B, de Graauw M. TNF- a -mediated NF- k B survival signaling impairment by cisplatin enhances JNK activation allowing synergistic apoptosis of renal proximal tubular cells. Biochem Pharmacol. 2013;85(2):274–86. 10.1016/j.bcp.2012.10.012. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

