Estimating Gene Conversion Tract Length and Rate From PacBio HiFi Data
- PMID: 39982809
- PMCID: PMC11844249
- DOI: 10.1093/molbev/msaf019
Estimating Gene Conversion Tract Length and Rate From PacBio HiFi Data
Abstract
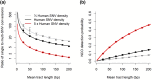
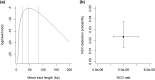
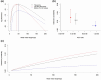
Gene conversions are broadly defined as the transfer of genetic material from a "donor" to an "acceptor" sequence and can happen both in meiosis and mitosis. They are a subset of noncrossover (NCO) events and, like crossover (CO) events, gene conversion can generate new combinations of alleles and counteract mutation load by reverting germline mutations through GC-biased gene conversion. Estimating gene conversion rate and the distribution of gene conversion tract lengths remains challenging. We present a new method for estimating tract length, rate, and detection probability of NCO events directly in HiFi PacBio long read data. The method can be used to make inference from sequencing of gametes from a single individual. The method is unbiased even under low single nucleotide variant (SNV) densities and does not necessitate any demographic or evolutionary assumptions. We test the accuracy and robustness of our method using simulated datasets where we vary length of tracts, number of tracts, the genomic SNV density, and levels of correlation between SNV density and NCO event position. Our simulations show that under low SNV densities, like those found in humans, only a minute fraction (∼2%) of NCO events are expected to become visible as gene conversions by moving at least 1 SNV. We finally illustrate our method by applying it to PacBio sequencing data from human sperm.
Keywords: gene conversion; genome evolution; genomics methods; noncrossover; recombination.
© The Author(s) 2025. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest: None
Figures


References
-
- Arndt PF, Massip F, Sheinman M. An analytical derivation of the distribution of distances between heterozygous sites in diploid species to efficiently infer demographic history. bioRxiv. 2023. 10.1101/2023.09.20.558510, 17 January 2025, preprint: not peer reviewed. - DOI
-
- Barroso GV, Dutheil JY. The landscape of nucleotide diversity in Drosophila melanogaster is shaped by mutation rate variation. Peer Community J. 2023:3:3–12. 10.24072/pcjournal.267. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

