An efficient, non-viral arrayed CRISPR screening platform for iPSC-derived myeloid and microglia models
- PMID: 39983727
- PMCID: PMC11960525
- DOI: 10.1016/j.stemcr.2025.102420
An efficient, non-viral arrayed CRISPR screening platform for iPSC-derived myeloid and microglia models
Abstract
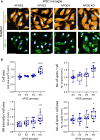
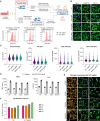
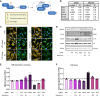
Here, we developed a CRISPR-Cas9 arrayed screen to investigate lipid handling pathways in human induced pluripotent stem cell (iPSC)-derived microglia. We established a robust method for the nucleofection of CRISPR-Cas9 ribonucleoprotein complexes into iPSC-derived myeloid cells, enabling genetic perturbations. Using this approach, we performed a targeted screen to identify key regulators of lipid droplet formation dependent on Apolipoprotein E (APOE). We identify the Mammalian Target of Rapamycin Complex 1 (mTORC1) signaling pathway as a critical modulator of lipid storage in both APOE3 and APOE knockout microglia. This study is a proof of concept underscoring the utility of CRISPR-Cas9 technology in elucidating the molecular pathways of lipid dysregulation associated with Alzheimer's disease and neuroinflammation.
Keywords: APOE; CRISPR-Cas9 gene editing; arrayed genetic screening; iPSC-derived microglia; lipid accumulation; lipid droplet screen; lipid metabolism; lipid regulation; lysosome; mTORC1.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests F.W., N.M., L.T., E.S.M., L.C., F.R., and R.J. are employees of F. Hoffman-La Roche Ltd. S.M., A.S.G.L., and A.M. were employees of F. Hoffman-La Roche Ltd at the time of the study. The design, research conduct, and financial support for this study were provided by F. Hoffman-La Roche Ltd. F.R. and R.J. are shareholders of F. Hoffman-La Roche Ltd. M.K. is a co-scientific founder of Montara Therapeutics; serves on the scientific advisory boards of Engine Biosciences, Casma Therapeutics, Alector, Montara Therapeutics, and Neurocrine Biosciences; and is an advisor to Modulo Bio and Recursion Therapeutics. M.K. is an inventor on the US Patent 11,254,933 related to CRISPRi and CRISPRa screening.
Figures

References
-
- Drager N.M., Sattler S.M., Huang C.T., Teter O.M., Leng K., Hashemi S.H., Hong J., Aviles G., Clelland C.D., Zhan L., et al. A CRISPRi/a platform in human iPSC-derived microglia uncovers regulators of disease states. Nat. Neurosci. 2022;25:1149–1162. doi: 10.1038/s41593-022-01131-4. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

