Structural and functional characterization of the extended-diKH domain from the antiviral endoribonuclease KHNYN
- PMID: 39984050
- PMCID: PMC11997328
- DOI: 10.1016/j.jbc.2025.108336
Structural and functional characterization of the extended-diKH domain from the antiviral endoribonuclease KHNYN
Abstract
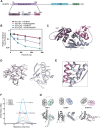
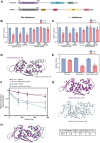
Zinc finger antiviral protein (ZAP) binds CpG dinucleotides in viral RNA and targets them for decay. ZAP interacts with several cofactors to form the ZAP antiviral system, including KHNYN, a multidomain endoribonuclease required for ZAP-mediated RNA decay. However, it is unclear how the individual domains in KHNYN contribute to its activity. Here, we demonstrate that the KHNYN amino-terminal extended-diKH (ex-diKH) domain is required for antiviral activity and present its crystal structure. The structure belongs to a rare group of KH-containing domains, characterized by a noncanonical arrangement between two type 1 KH modules, with an additional helical bundle. N4BP1 is a KHNYN paralog with an ex-diKH domain that functionally complements the KHNYN ex-diKH domain. Interestingly, the ex-diKH domain structure is present in N4BP1-like proteins in lancelets, which are basal chordates, indicating that it is evolutionarily ancient. While many KH domains demonstrate RNA binding activity, biolayer interferometry and electrophoretic mobility shift assays indicate that the KHNYN ex-diKH domain does not bind RNA. Furthermore, residues required for canonical KH domains to bind RNA are not required for KHNYN antiviral activity. By contrast, an inter-KH domain cleft in KHNYN is a potential protein-protein interaction site, and mutations that eliminate arginine salt bridges at the edge of this cleft decrease KHNYN antiviral activity. This suggests that this domain could be a binding site for an unknown KHNYN cofactor.
Keywords: CpG; HIV; KHNYN; RNA turnover; RNA virus; ZAP; ZC3HAV1; innate immunity; structure-function; viral replication.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Gao G., Guo X., Goff S.P. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297:1703–1706. - PubMed
-
- Ficarelli M., Neil S.J.D., Swanson C.M. Targeted restriction of viral gene expression and replication by the ZAP antiviral system. Annu. Rev. Virol. 2021;8:265–283. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

